Iron »
PDB 8sqp-8u19 »
8tgb »
Iron in PDB 8tgb: Crystal Structure of Root Lateral Formation Protein (Rlf) B5-Domain From Oryza Sativa
Protein crystallography data
The structure of Crystal Structure of Root Lateral Formation Protein (Rlf) B5-Domain From Oryza Sativa, PDB code: 8tgb
was solved by
S.Lovell,
M.M.Kashipathy,
K.P.Battaile,
D.R.Benson,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 41.25 / 1.55 |
| Space group | P 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 58.951, 26.138, 82.873, 90, 95.5, 90 |
| R / Rfree (%) | 17.5 / 20 |
Iron Binding Sites:
The binding sites of Iron atom in the Crystal Structure of Root Lateral Formation Protein (Rlf) B5-Domain From Oryza Sativa
(pdb code 8tgb). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 2 binding sites of Iron where determined in the Crystal Structure of Root Lateral Formation Protein (Rlf) B5-Domain From Oryza Sativa, PDB code: 8tgb:
Jump to Iron binding site number: 1; 2;
In total 2 binding sites of Iron where determined in the Crystal Structure of Root Lateral Formation Protein (Rlf) B5-Domain From Oryza Sativa, PDB code: 8tgb:
Jump to Iron binding site number: 1; 2;
Iron binding site 1 out of 2 in 8tgb
Go back to
Iron binding site 1 out
of 2 in the Crystal Structure of Root Lateral Formation Protein (Rlf) B5-Domain From Oryza Sativa
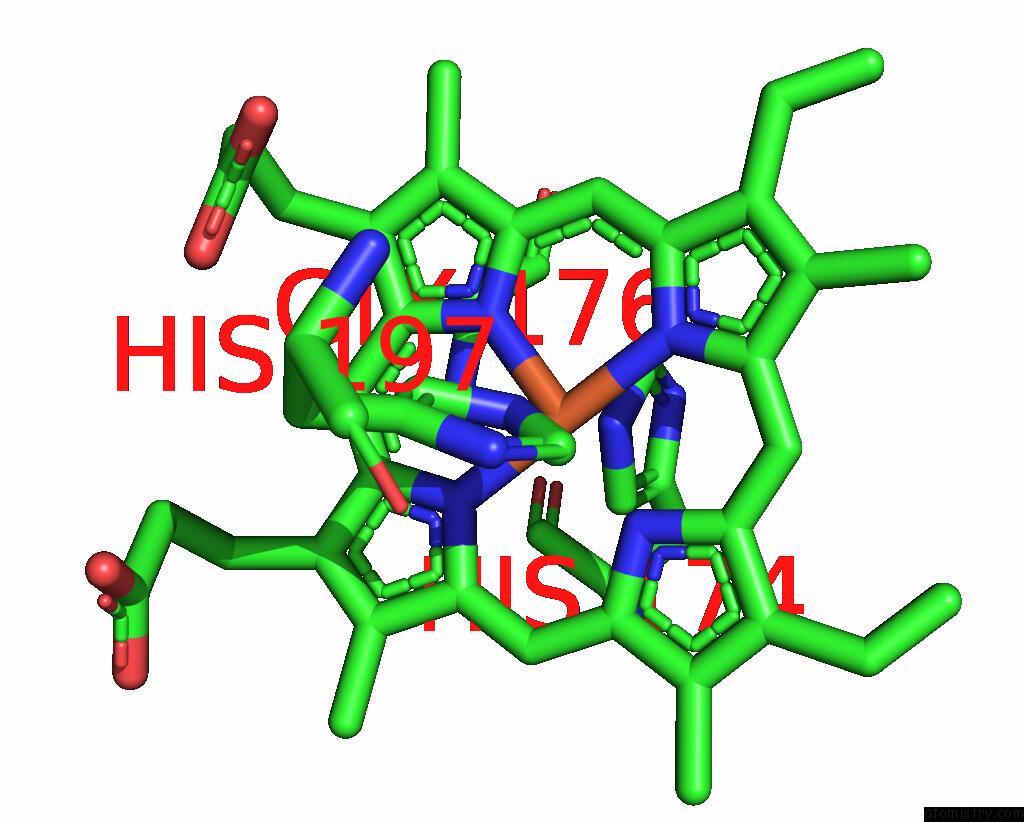
Mono view
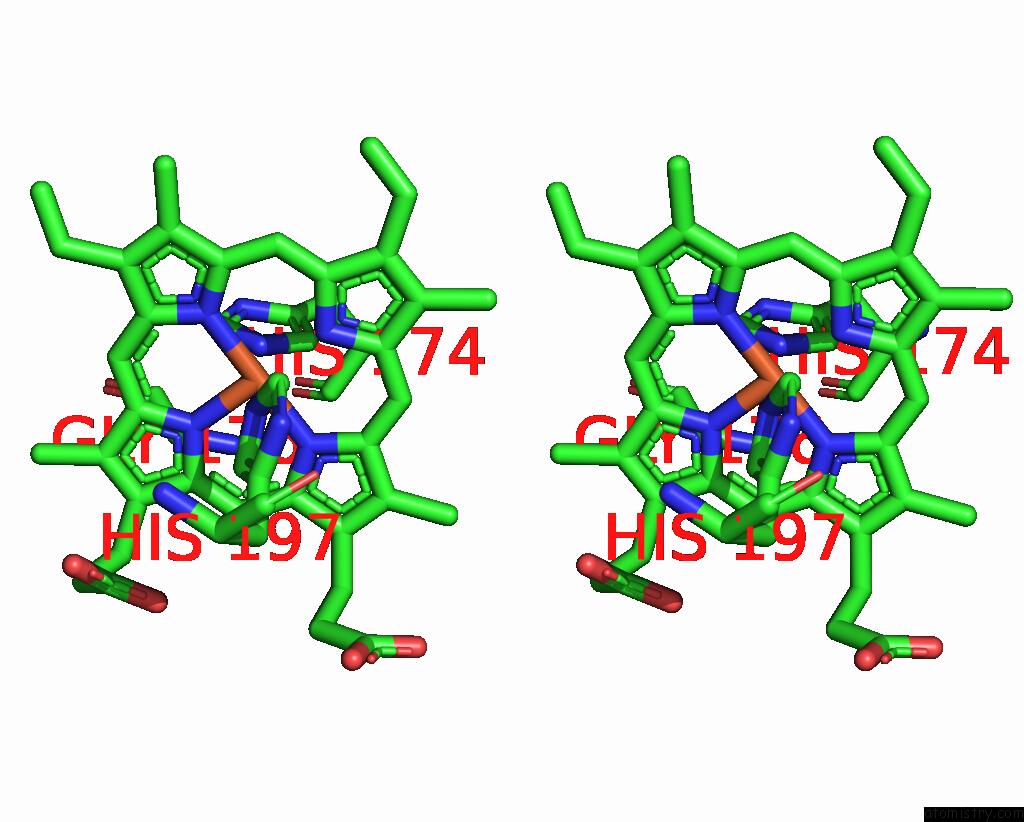
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Crystal Structure of Root Lateral Formation Protein (Rlf) B5-Domain From Oryza Sativa within 5.0Å range:
|
Iron binding site 2 out of 2 in 8tgb
Go back to
Iron binding site 2 out
of 2 in the Crystal Structure of Root Lateral Formation Protein (Rlf) B5-Domain From Oryza Sativa
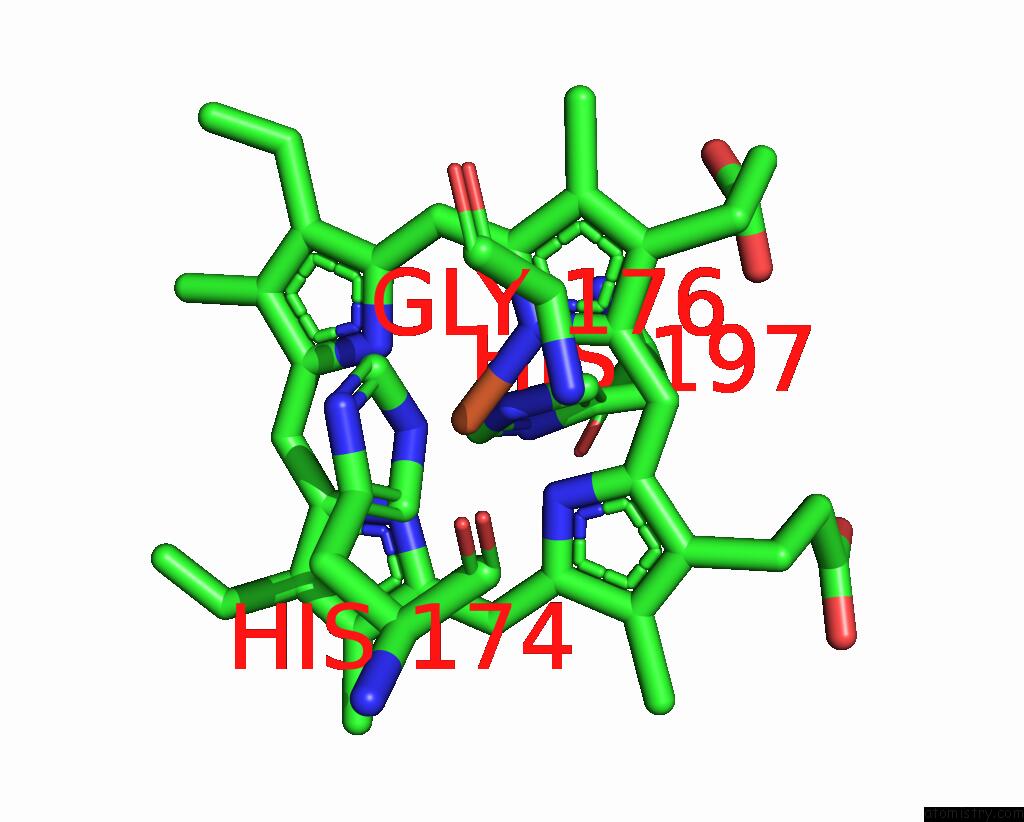
Mono view
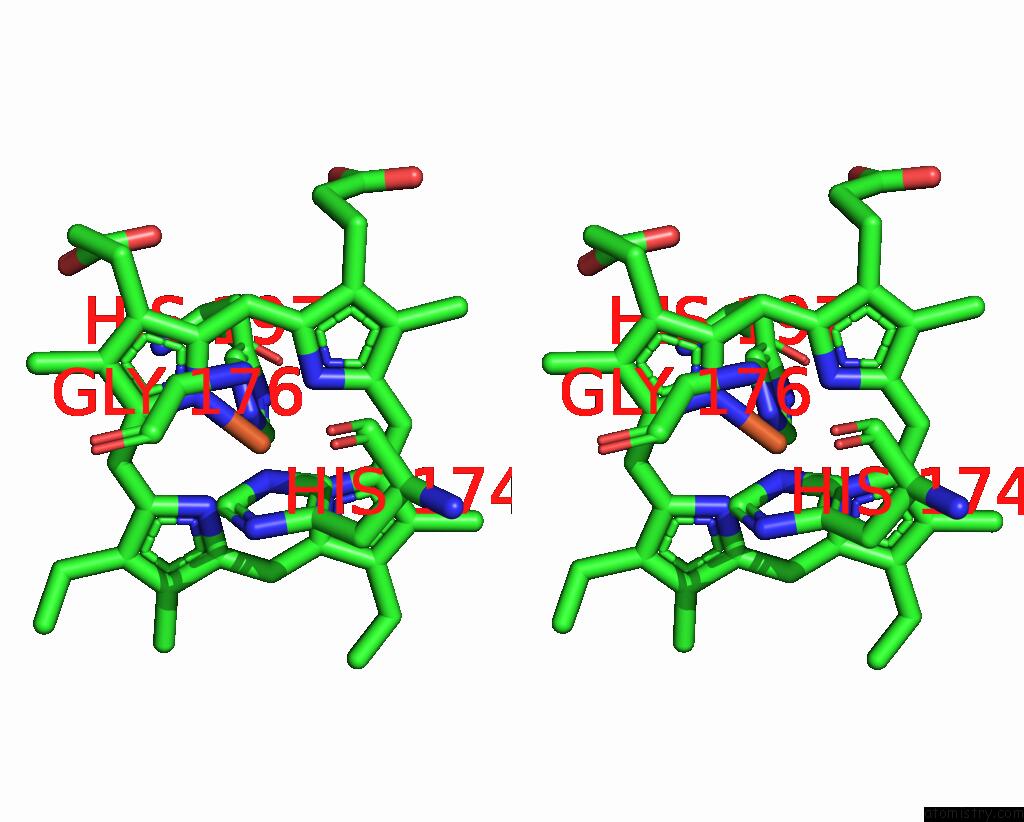
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Crystal Structure of Root Lateral Formation Protein (Rlf) B5-Domain From Oryza Sativa within 5.0Å range:
|
Reference:
D.R.Benson,
B.Deng,
M.M.Kashipathy,
S.Lovell,
K.P.Battaile,
A.Cooper,
P.Gao,
A.W.Fenton,
H.Zhu.
The N-Terminal Intrinsically Disordered Region of NCB5OR Docks with the Cytochrome B 5 Core to Form A Helical Motif That Is of Ancient Origin. Proteins 2023.
ISSN: ESSN 1097-0134
PubMed: 38041394
DOI: 10.1002/PROT.26647
Page generated: Thu Aug 7 23:43:35 2025
ISSN: ESSN 1097-0134
PubMed: 38041394
DOI: 10.1002/PROT.26647
Last articles
Fe in 9C1ZFe in 9C1Y
Fe in 9C0R
Fe in 9C0Q
Fe in 9BT4
Fe in 9BUO
Fe in 9BWU
Fe in 9BTS
Fe in 9BW8
Fe in 9BP5