Iron »
PDB 1jl6-1k2o »
1jmz »
Iron in PDB 1jmz: Crystal Structure of A Quinohemoprotein Amine Dehydrogenase From Pseudomonas Putida with Inhibitor
Protein crystallography data
The structure of Crystal Structure of A Quinohemoprotein Amine Dehydrogenase From Pseudomonas Putida with Inhibitor, PDB code: 1jmz
was solved by
A.Satoh,
I.Miyahara,
K.Hirotsu,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.00 / 2.00 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 167.210, 92.370, 79.300, 90.00, 112.00, 90.00 |
| R / Rfree (%) | 21.5 / 25.1 |
Other elements in 1jmz:
The structure of Crystal Structure of A Quinohemoprotein Amine Dehydrogenase From Pseudomonas Putida with Inhibitor also contains other interesting chemical elements:
| Nickel | (Ni) | 1 atom |
Iron Binding Sites:
The binding sites of Iron atom in the Crystal Structure of A Quinohemoprotein Amine Dehydrogenase From Pseudomonas Putida with Inhibitor
(pdb code 1jmz). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 2 binding sites of Iron where determined in the Crystal Structure of A Quinohemoprotein Amine Dehydrogenase From Pseudomonas Putida with Inhibitor, PDB code: 1jmz:
Jump to Iron binding site number: 1; 2;
In total 2 binding sites of Iron where determined in the Crystal Structure of A Quinohemoprotein Amine Dehydrogenase From Pseudomonas Putida with Inhibitor, PDB code: 1jmz:
Jump to Iron binding site number: 1; 2;
Iron binding site 1 out of 2 in 1jmz
Go back to
Iron binding site 1 out
of 2 in the Crystal Structure of A Quinohemoprotein Amine Dehydrogenase From Pseudomonas Putida with Inhibitor
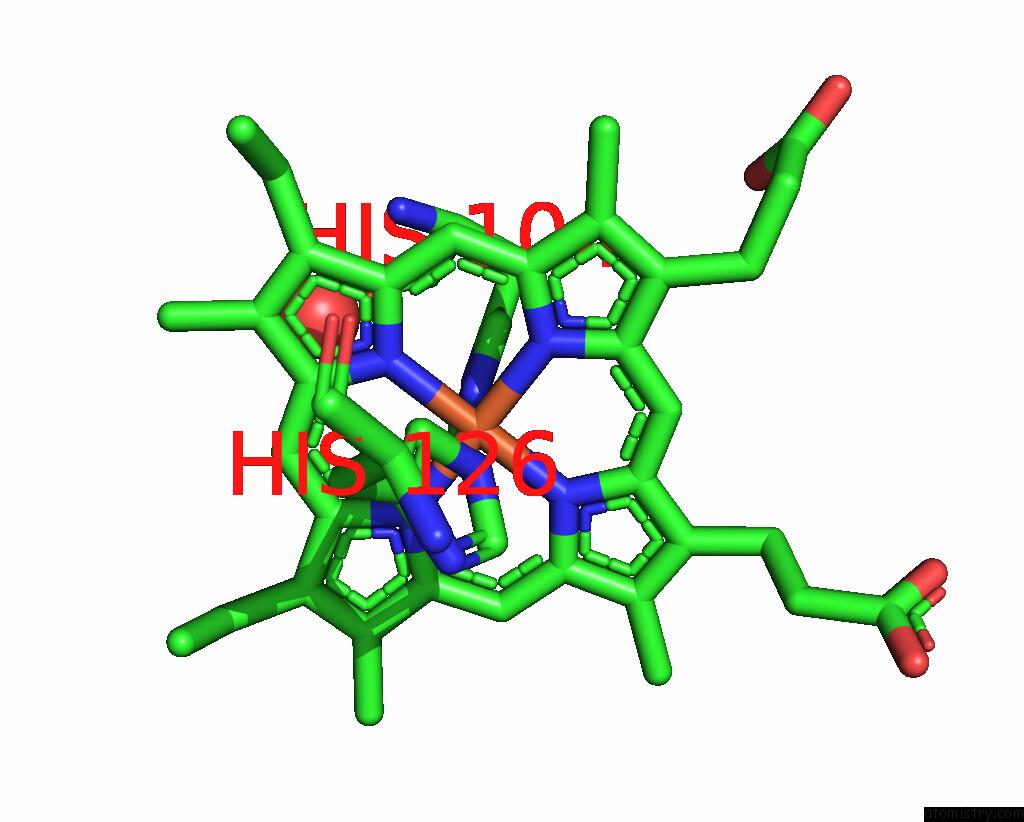
Mono view
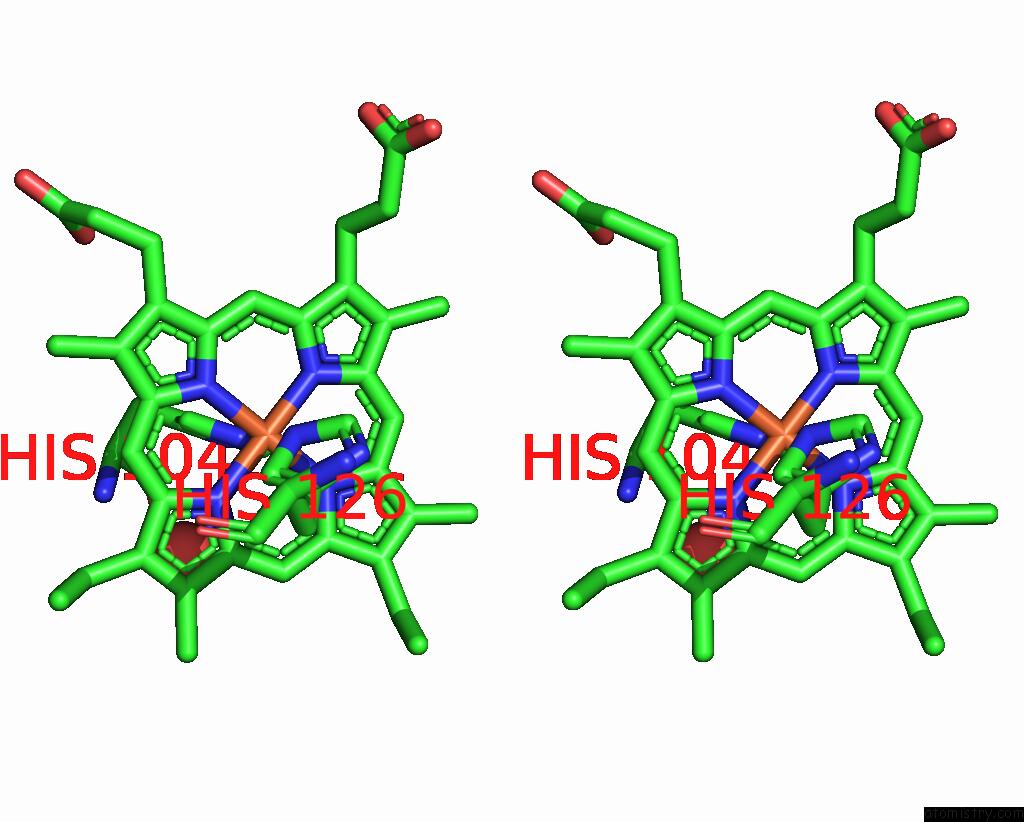
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Crystal Structure of A Quinohemoprotein Amine Dehydrogenase From Pseudomonas Putida with Inhibitor within 5.0Å range:
|
Iron binding site 2 out of 2 in 1jmz
Go back to
Iron binding site 2 out
of 2 in the Crystal Structure of A Quinohemoprotein Amine Dehydrogenase From Pseudomonas Putida with Inhibitor
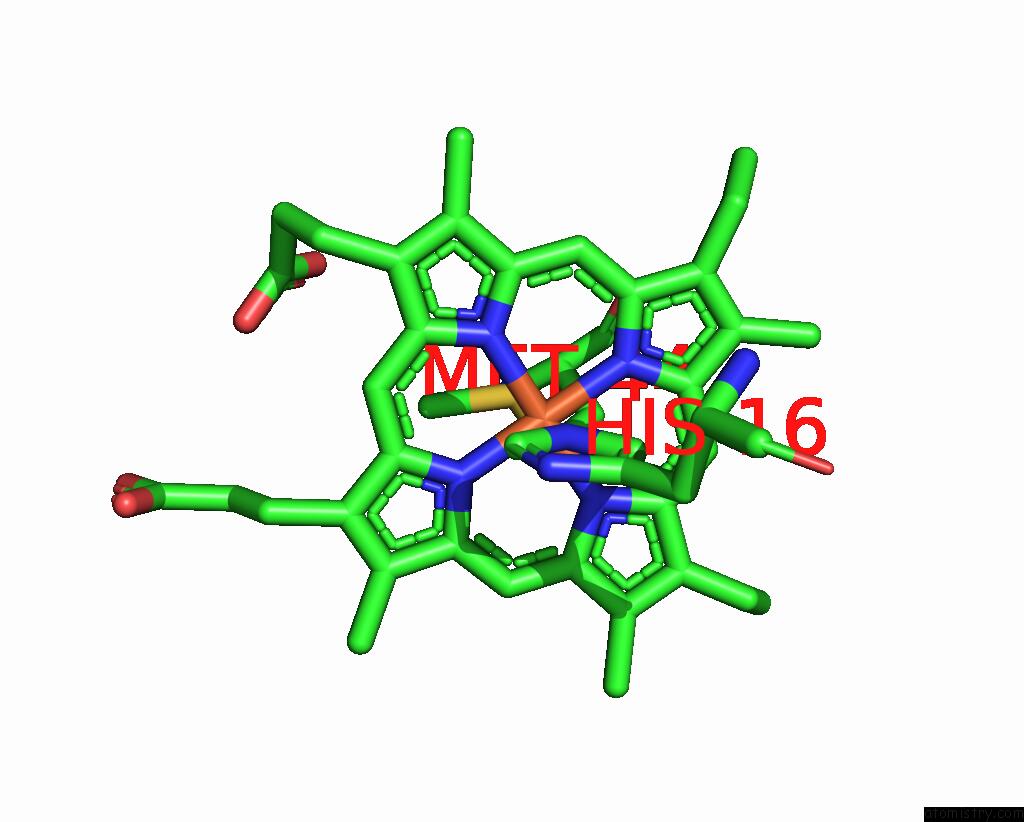
Mono view
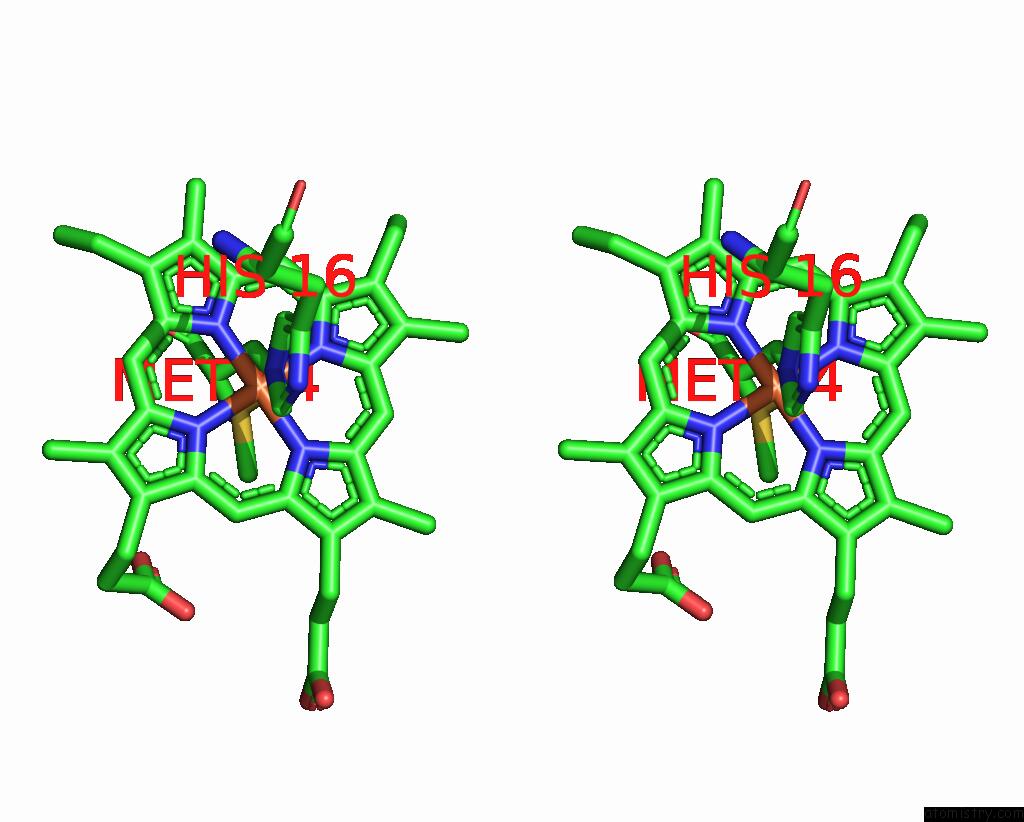
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Crystal Structure of A Quinohemoprotein Amine Dehydrogenase From Pseudomonas Putida with Inhibitor within 5.0Å range:
|
Reference:
A.Satoh,
J.K.Kim,
I.Miyahara,
B.Devreese,
I.Vandenberghe,
A.Hacisalihoglu,
T.Okajima,
S.Kuroda,
O.Adachi,
J.A.Duine,
J.Van Beeumen,
K.Tanizawa,
K.Hirotsu.
Crystal Structure of Quinohemoprotein Amine Dehydrogenase From Pseudomonas Putida. Identification of A Novel Quinone Cofactor Encaged By Multiple Thioether Cross-Bridges. J.Biol.Chem. V. 277 2830 2002.
ISSN: ISSN 0021-9258
PubMed: 11704672
DOI: 10.1074/JBC.M109090200
Page generated: Sat Aug 3 08:35:21 2024
ISSN: ISSN 0021-9258
PubMed: 11704672
DOI: 10.1074/JBC.M109090200
Last articles
F in 9MSQF in 9MEL
F in 9LRJ
F in 9M3P
F in 9LJ5
F in 9JVW
F in 9LJ1
F in 9JJV
F in 9JIP
F in 9JI1