Iron »
PDB 1lfk-1lr6 »
1lkm »
Iron in PDB 1lkm: Crystal Structure of Desulfovibrio Vulgaris Rubrerythrin All-Iron(III) Form
Protein crystallography data
The structure of Crystal Structure of Desulfovibrio Vulgaris Rubrerythrin All-Iron(III) Form, PDB code: 1lkm
was solved by
S.Jin,
D.M.Kurtz Jr.,
Z.J.Liu,
J.Rose,
B.C.Wang,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 25.95 / 1.69 |
| Space group | I 2 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 48.877, 80.609, 100.084, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19 / 21 |
Iron Binding Sites:
The binding sites of Iron atom in the Crystal Structure of Desulfovibrio Vulgaris Rubrerythrin All-Iron(III) Form
(pdb code 1lkm). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 3 binding sites of Iron where determined in the Crystal Structure of Desulfovibrio Vulgaris Rubrerythrin All-Iron(III) Form, PDB code: 1lkm:
Jump to Iron binding site number: 1; 2; 3;
In total 3 binding sites of Iron where determined in the Crystal Structure of Desulfovibrio Vulgaris Rubrerythrin All-Iron(III) Form, PDB code: 1lkm:
Jump to Iron binding site number: 1; 2; 3;
Iron binding site 1 out of 3 in 1lkm
Go back to
Iron binding site 1 out
of 3 in the Crystal Structure of Desulfovibrio Vulgaris Rubrerythrin All-Iron(III) Form
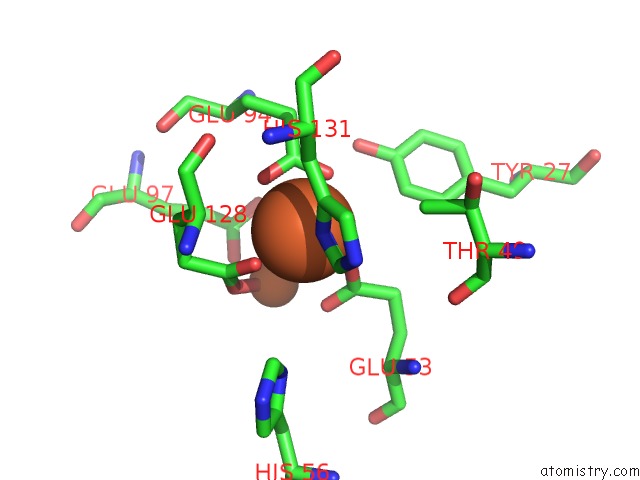
Mono view
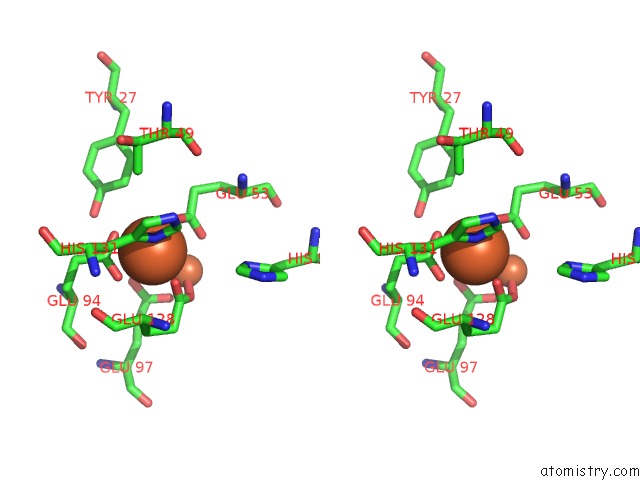
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Crystal Structure of Desulfovibrio Vulgaris Rubrerythrin All-Iron(III) Form within 5.0Å range:
|
Iron binding site 2 out of 3 in 1lkm
Go back to
Iron binding site 2 out
of 3 in the Crystal Structure of Desulfovibrio Vulgaris Rubrerythrin All-Iron(III) Form
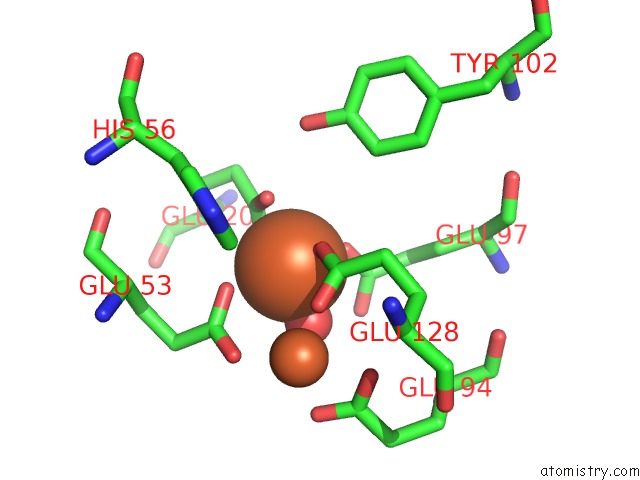
Mono view
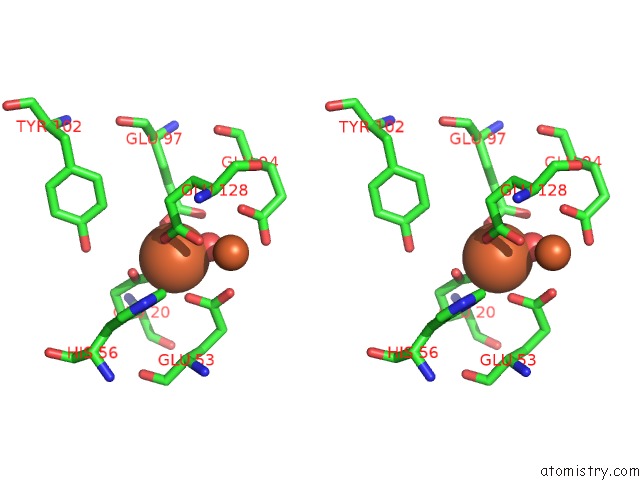
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Crystal Structure of Desulfovibrio Vulgaris Rubrerythrin All-Iron(III) Form within 5.0Å range:
|
Iron binding site 3 out of 3 in 1lkm
Go back to
Iron binding site 3 out
of 3 in the Crystal Structure of Desulfovibrio Vulgaris Rubrerythrin All-Iron(III) Form
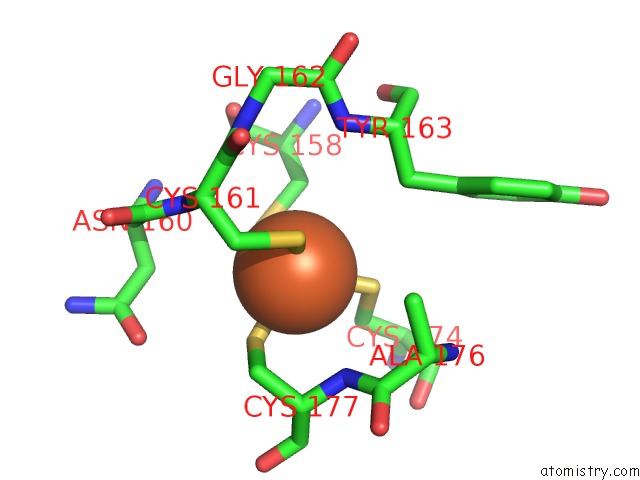
Mono view
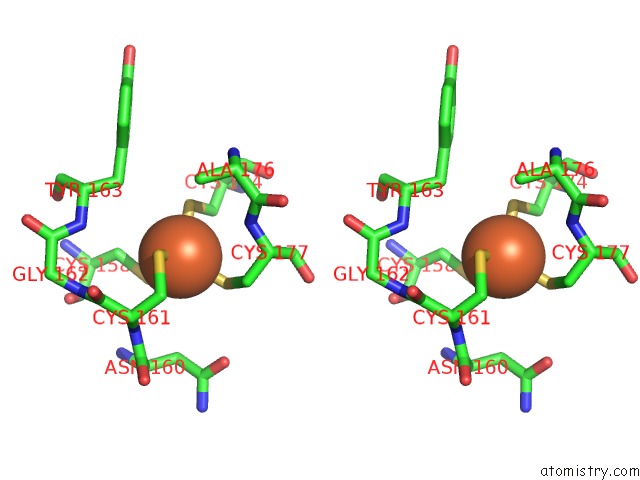
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 3 of Crystal Structure of Desulfovibrio Vulgaris Rubrerythrin All-Iron(III) Form within 5.0Å range:
|
Reference:
S.Jin,
D.M.Kurtz Jr.,
Z.J.Liu,
J.Rose,
B.C.Wang.
X-Ray Crystal Structures of Reduced Rubrerythrin and Its Azide Adduct: A Structure-Based Mechanism For A Non-Heme Diiron Peroxidase J.Am.Chem.Soc. V. 124 9845 2002.
ISSN: ISSN 0002-7863
PubMed: 12175244
DOI: 10.1021/JA026587U
Page generated: Wed Jul 16 17:26:11 2025
ISSN: ISSN 0002-7863
PubMed: 12175244
DOI: 10.1021/JA026587U
Last articles
F in 9VCLF in 9VCK
F in 9JHS
F in 9JF9
F in 9CPK
Cu in 9OH6
Cu in 9OH7
Cu in 9VQM
Cl in 9QM5
Cl in 9RM2