Iron »
PDB 1nml-1o1i »
1nnt »
Iron in PDB 1nnt: Structural Evidence For A pH-Sensitive Di-Lysine Trigger in the Hen Ovotransferrin N-Lobe: Implications For Transferrin Iron Release
Protein crystallography data
The structure of Structural Evidence For A pH-Sensitive Di-Lysine Trigger in the Hen Ovotransferrin N-Lobe: Implications For Transferrin Iron Release, PDB code: 1nnt
was solved by
J.C.Dewan,
B.Mikami,
J.C.Sacchettini,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | N/A / 2.30 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 46.940, 91.790, 75.820, 90.00, 90.00, 90.00 |
| R / Rfree (%) | n/a / n/a |
Iron Binding Sites:
The binding sites of Iron atom in the Structural Evidence For A pH-Sensitive Di-Lysine Trigger in the Hen Ovotransferrin N-Lobe: Implications For Transferrin Iron Release
(pdb code 1nnt). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total only one binding site of Iron was determined in the Structural Evidence For A pH-Sensitive Di-Lysine Trigger in the Hen Ovotransferrin N-Lobe: Implications For Transferrin Iron Release, PDB code: 1nnt:
In total only one binding site of Iron was determined in the Structural Evidence For A pH-Sensitive Di-Lysine Trigger in the Hen Ovotransferrin N-Lobe: Implications For Transferrin Iron Release, PDB code: 1nnt:
Iron binding site 1 out of 1 in 1nnt
Go back to
Iron binding site 1 out
of 1 in the Structural Evidence For A pH-Sensitive Di-Lysine Trigger in the Hen Ovotransferrin N-Lobe: Implications For Transferrin Iron Release
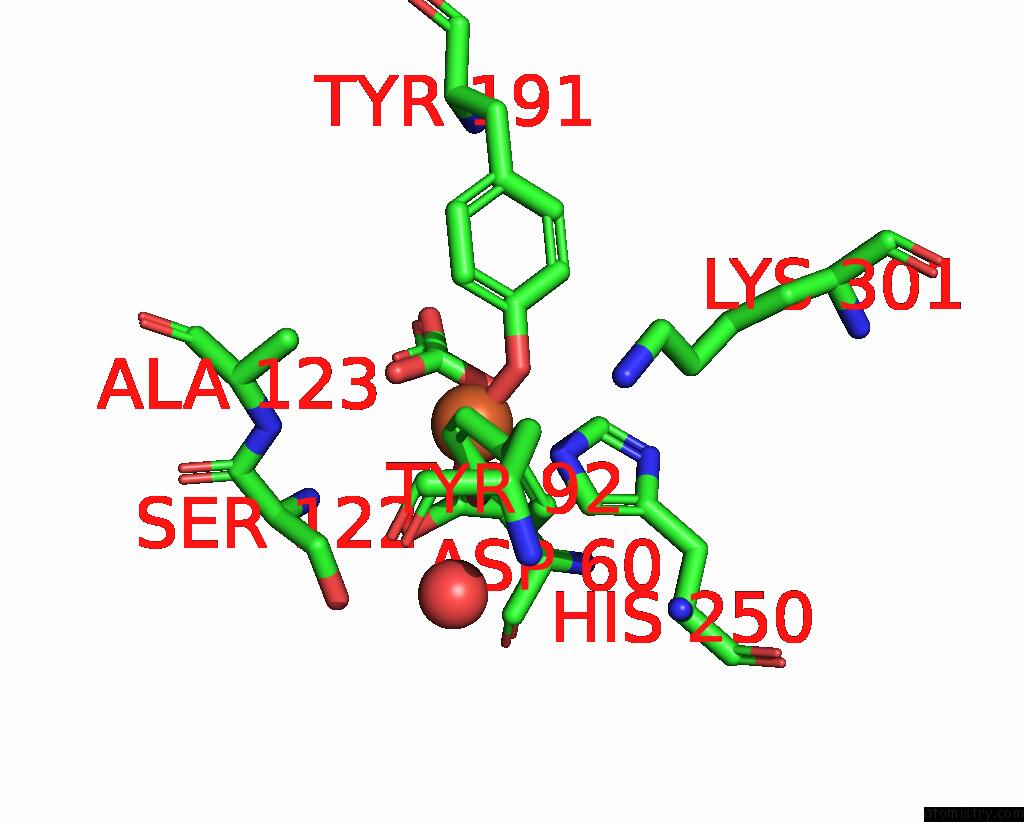
Mono view
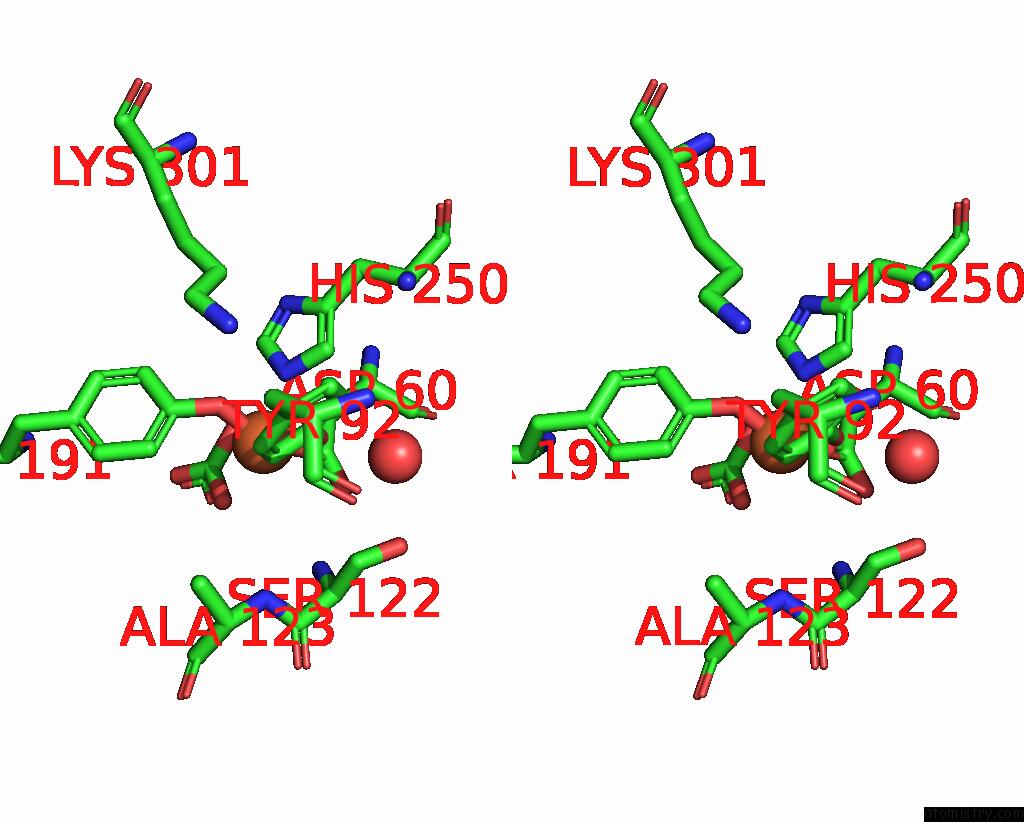
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Structural Evidence For A pH-Sensitive Di-Lysine Trigger in the Hen Ovotransferrin N-Lobe: Implications For Transferrin Iron Release within 5.0Å range:
|
Reference:
J.C.Dewan,
B.Mikami,
M.Hirose,
J.C.Sacchettini.
Structural Evidence For A pH-Sensitive Dilysine Trigger in the Hen Ovotransferrin N-Lobe: Implications For Transferrin Iron Release. Biochemistry V. 32 11963 1993.
ISSN: ISSN 0006-2960
PubMed: 8218271
DOI: 10.1021/BI00096A004
Page generated: Wed Jul 16 18:49:59 2025
ISSN: ISSN 0006-2960
PubMed: 8218271
DOI: 10.1021/BI00096A004
Last articles
Mg in 3J7HMg in 3J7I
Mg in 3IVK
Mg in 3J6E
Mg in 3J6G
Mg in 3J6P
Mg in 3J6H
Mg in 3J1F
Mg in 3J6F
Mg in 3J5V