Iron »
PDB 1sdl-1stq »
1smw »
Iron in PDB 1smw: Crystal Structure of Cp Rd L41A Mutant in Reduced State 2 (Soaked)
Protein crystallography data
The structure of Crystal Structure of Cp Rd L41A Mutant in Reduced State 2 (Soaked), PDB code: 1smw
was solved by
I.Y.Park,
B.Youn,
J.L.Harley,
M.K.Eidsness,
E.Smith,
T.Ichiye,
C.Kang,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.00 / 1.38 |
| Space group | H 3 |
| Cell size a, b, c (Å), α, β, γ (°) | 63.042, 63.042, 32.691, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 18.3 / 20 |
Iron Binding Sites:
The binding sites of Iron atom in the Crystal Structure of Cp Rd L41A Mutant in Reduced State 2 (Soaked)
(pdb code 1smw). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total only one binding site of Iron was determined in the Crystal Structure of Cp Rd L41A Mutant in Reduced State 2 (Soaked), PDB code: 1smw:
In total only one binding site of Iron was determined in the Crystal Structure of Cp Rd L41A Mutant in Reduced State 2 (Soaked), PDB code: 1smw:
Iron binding site 1 out of 1 in 1smw
Go back to
Iron binding site 1 out
of 1 in the Crystal Structure of Cp Rd L41A Mutant in Reduced State 2 (Soaked)
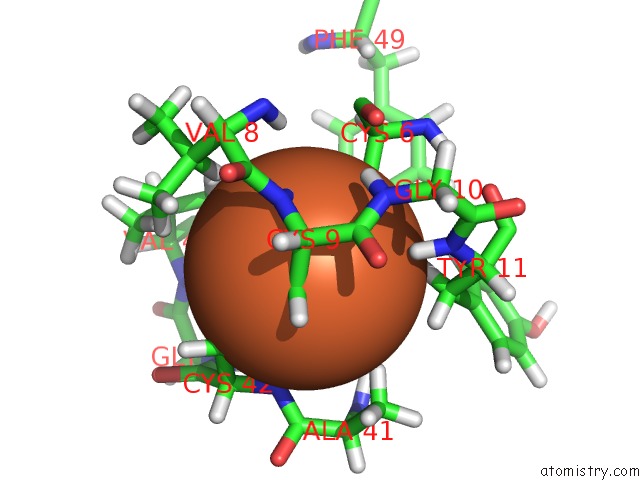
Mono view
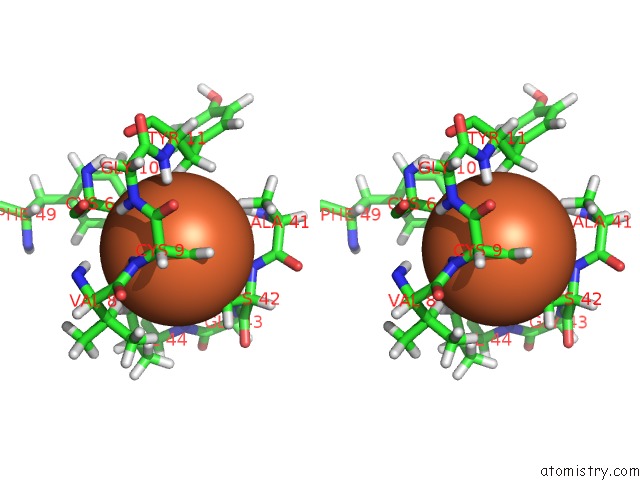
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Crystal Structure of Cp Rd L41A Mutant in Reduced State 2 (Soaked) within 5.0Å range:
|
Reference:
I.Y.Park,
B.Youn,
J.L.Harley,
M.K.Eidsness,
E.Smith,
T.Ichiye,
C.Kang.
The Unique Hydrogen Bonded Water in the Reduced Form of Clostridium Pasteurianum Rubredoxin and Its Possible Role in Electron Transfer J.Biol.Inorg.Chem. V. 9 423 2004.
ISSN: ISSN 0949-8257
PubMed: 15067525
DOI: 10.1007/S00775-004-0542-3
Page generated: Wed Jul 16 20:42:38 2025
ISSN: ISSN 0949-8257
PubMed: 15067525
DOI: 10.1007/S00775-004-0542-3
Last articles
Zn in 1KH7Zn in 1KH5
Zn in 1KH4
Zn in 1KFV
Zn in 1KFS
Zn in 1KEV
Zn in 1KFI
Zn in 1KBP
Zn in 1KEQ
Zn in 1KEI