Iron »
PDB 1vrb-1w9m »
1w92 »
Iron in PDB 1w92: The Structure of Carbomonoxy Murine Neuroglobin Reveals A Heme- Sliding Mechanism For Affinity Regulation
Protein crystallography data
The structure of The Structure of Carbomonoxy Murine Neuroglobin Reveals A Heme- Sliding Mechanism For Affinity Regulation, PDB code: 1w92
was solved by
B.Vallone,
K.Nienhaus,
A.Matthes,
M.Brunori,
G.U.Nienhaus,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 15.00 / 1.70 |
| Space group | H 3 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 88.373, 88.373, 110.983, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 26 / 22 |
Iron Binding Sites:
The binding sites of Iron atom in the The Structure of Carbomonoxy Murine Neuroglobin Reveals A Heme- Sliding Mechanism For Affinity Regulation
(pdb code 1w92). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total only one binding site of Iron was determined in the The Structure of Carbomonoxy Murine Neuroglobin Reveals A Heme- Sliding Mechanism For Affinity Regulation, PDB code: 1w92:
In total only one binding site of Iron was determined in the The Structure of Carbomonoxy Murine Neuroglobin Reveals A Heme- Sliding Mechanism For Affinity Regulation, PDB code: 1w92:
Iron binding site 1 out of 1 in 1w92
Go back to
Iron binding site 1 out
of 1 in the The Structure of Carbomonoxy Murine Neuroglobin Reveals A Heme- Sliding Mechanism For Affinity Regulation
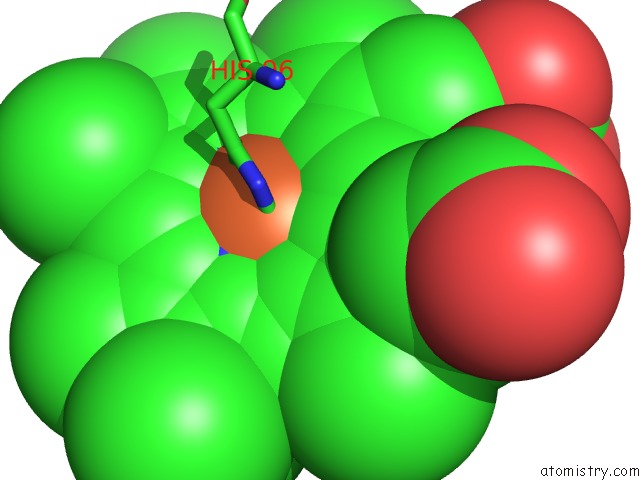
Mono view
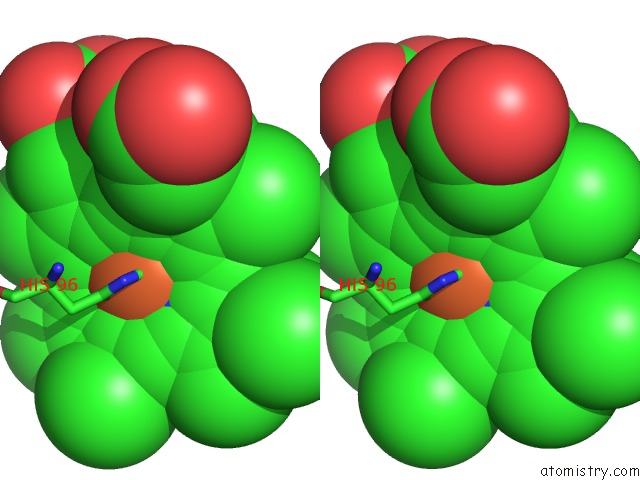
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of The Structure of Carbomonoxy Murine Neuroglobin Reveals A Heme- Sliding Mechanism For Affinity Regulation within 5.0Å range:
|
Reference:
B.Vallone,
K.Nienhaus,
A.Matthes,
M.Brunori,
G.U.Nienhaus.
The Structure of Carbonmonoxy Neuroglobin Reveals A Heme-Sliding Mechanism For Control of Ligand Affinity Proc.Natl.Acad.Sci.Usa V. 101 17351 2004.
ISSN: ISSN 0027-8424
PubMed: 15548613
DOI: 10.1073/PNAS.0407633101
Page generated: Wed Jul 16 21:48:29 2025
ISSN: ISSN 0027-8424
PubMed: 15548613
DOI: 10.1073/PNAS.0407633101
Last articles
Zn in 1LBUZn in 1LAN
Zn in 1LBA
Zn in 1LAP
Zn in 1LAM
Zn in 1L9H
Zn in 1L9Y
Zn in 1L9Z
Zn in 1L8C
Zn in 1L9U