Iron »
PDB 2ibz-2j2f »
2iuf »
Iron in PDB 2iuf: The Structures of Penicillium Vitale Catalase: Resting State, Oxidised State (Compound I) and Complex with Aminotriazole
Enzymatic activity of The Structures of Penicillium Vitale Catalase: Resting State, Oxidised State (Compound I) and Complex with Aminotriazole
All present enzymatic activity of The Structures of Penicillium Vitale Catalase: Resting State, Oxidised State (Compound I) and Complex with Aminotriazole:
1.11.1.6;
1.11.1.6;
Protein crystallography data
The structure of The Structures of Penicillium Vitale Catalase: Resting State, Oxidised State (Compound I) and Complex with Aminotriazole, PDB code: 2iuf
was solved by
G.Murshudov,
A.Borovik,
A.Grebenko,
V.Barynin,
A.Vagin,
W.Melik-Adamyan,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 119.52 / 1.71 |
| Space group | P 31 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 142.440, 142.440, 132.230, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 12.9 / 15.4 |
Other elements in 2iuf:
The structure of The Structures of Penicillium Vitale Catalase: Resting State, Oxidised State (Compound I) and Complex with Aminotriazole also contains other interesting chemical elements:
| Calcium | (Ca) | 6 atoms |
Iron Binding Sites:
The binding sites of Iron atom in the The Structures of Penicillium Vitale Catalase: Resting State, Oxidised State (Compound I) and Complex with Aminotriazole
(pdb code 2iuf). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 2 binding sites of Iron where determined in the The Structures of Penicillium Vitale Catalase: Resting State, Oxidised State (Compound I) and Complex with Aminotriazole, PDB code: 2iuf:
Jump to Iron binding site number: 1; 2;
In total 2 binding sites of Iron where determined in the The Structures of Penicillium Vitale Catalase: Resting State, Oxidised State (Compound I) and Complex with Aminotriazole, PDB code: 2iuf:
Jump to Iron binding site number: 1; 2;
Iron binding site 1 out of 2 in 2iuf
Go back to
Iron binding site 1 out
of 2 in the The Structures of Penicillium Vitale Catalase: Resting State, Oxidised State (Compound I) and Complex with Aminotriazole
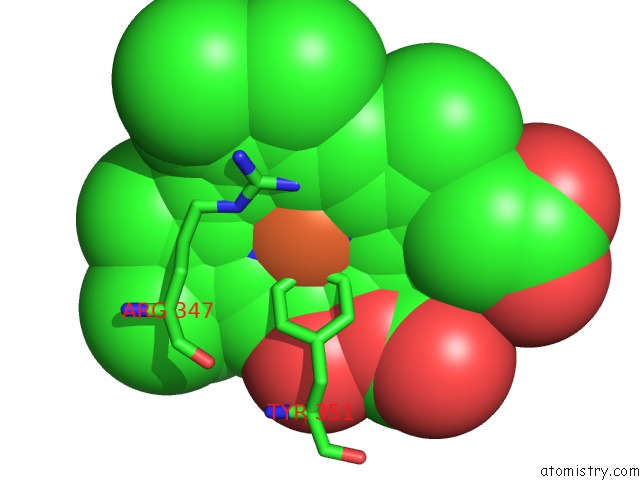
Mono view
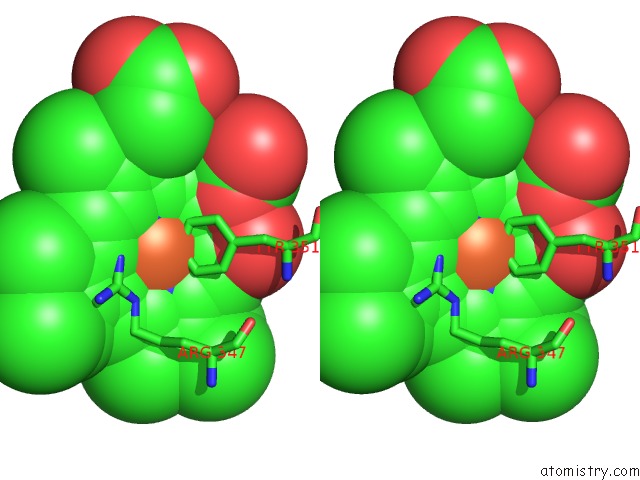
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of The Structures of Penicillium Vitale Catalase: Resting State, Oxidised State (Compound I) and Complex with Aminotriazole within 5.0Å range:
|
Iron binding site 2 out of 2 in 2iuf
Go back to
Iron binding site 2 out
of 2 in the The Structures of Penicillium Vitale Catalase: Resting State, Oxidised State (Compound I) and Complex with Aminotriazole
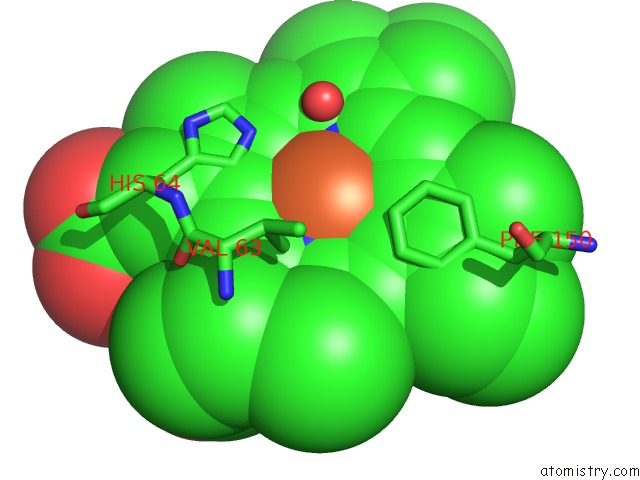
Mono view
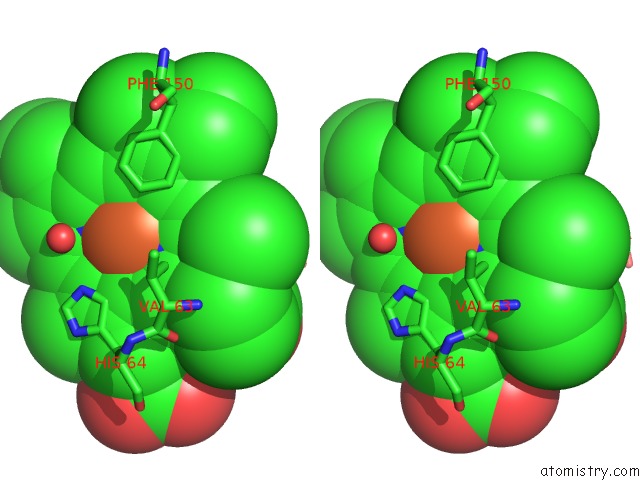
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of The Structures of Penicillium Vitale Catalase: Resting State, Oxidised State (Compound I) and Complex with Aminotriazole within 5.0Å range:
|
Reference:
M.Alfonso-Prieto,
A.Borovik,
X.Carpena,
G.Murshudov,
W.Melik-Adamyan,
I.Fita,
C.Rovira,
P.C.Loewen.
The Structures and Electronic Configuration of Compound I Intermediates of Helicobacter Pylori and Penicillium Vitale Catalases Determined By X-Ray Crystallography and Qm/Mm Density Functional Theory Calculations. J.Am.Chem.Soc. V. 129 4193 2007.
ISSN: ISSN 0002-7863
PubMed: 17358056
DOI: 10.1021/JA063660Y
Page generated: Thu Jul 17 02:19:39 2025
ISSN: ISSN 0002-7863
PubMed: 17358056
DOI: 10.1021/JA063660Y
Last articles
K in 3SRDK in 3SRK
K in 3SS6
K in 3SPJ
K in 3SPG
K in 3SPI
K in 3SPH
K in 3SPC
K in 3SBR
K in 3SBP