Iron »
PDB 2j2m-2ksu »
2jti »
Iron in PDB 2jti: Solution Structure of the Yeast Iso-1-Cytochrome C (T12A) : Yeast Cytochrome C Peroxidase Complex
Enzymatic activity of Solution Structure of the Yeast Iso-1-Cytochrome C (T12A) : Yeast Cytochrome C Peroxidase Complex
All present enzymatic activity of Solution Structure of the Yeast Iso-1-Cytochrome C (T12A) : Yeast Cytochrome C Peroxidase Complex:
1.11.1.5;
1.11.1.5;
Iron Binding Sites:
The binding sites of Iron atom in the Solution Structure of the Yeast Iso-1-Cytochrome C (T12A) : Yeast Cytochrome C Peroxidase Complex
(pdb code 2jti). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 2 binding sites of Iron where determined in the Solution Structure of the Yeast Iso-1-Cytochrome C (T12A) : Yeast Cytochrome C Peroxidase Complex, PDB code: 2jti:
Jump to Iron binding site number: 1; 2;
In total 2 binding sites of Iron where determined in the Solution Structure of the Yeast Iso-1-Cytochrome C (T12A) : Yeast Cytochrome C Peroxidase Complex, PDB code: 2jti:
Jump to Iron binding site number: 1; 2;
Iron binding site 1 out of 2 in 2jti
Go back to
Iron binding site 1 out
of 2 in the Solution Structure of the Yeast Iso-1-Cytochrome C (T12A) : Yeast Cytochrome C Peroxidase Complex
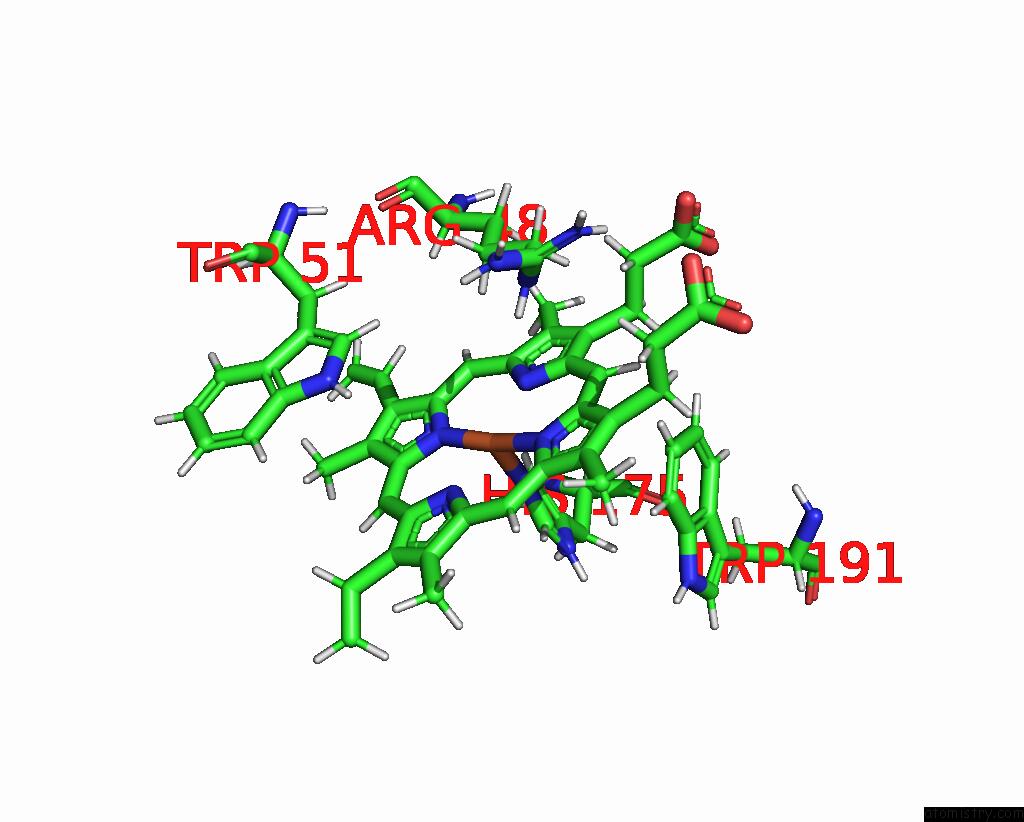
Mono view
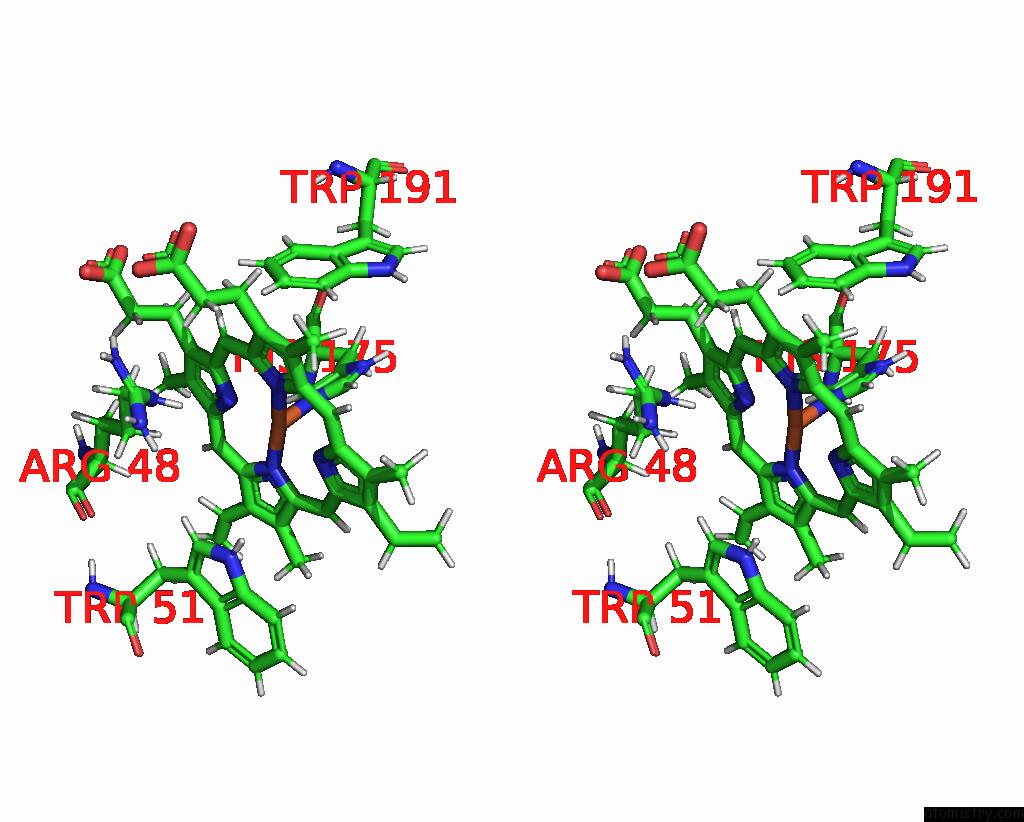
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Solution Structure of the Yeast Iso-1-Cytochrome C (T12A) : Yeast Cytochrome C Peroxidase Complex within 5.0Å range:
|
Iron binding site 2 out of 2 in 2jti
Go back to
Iron binding site 2 out
of 2 in the Solution Structure of the Yeast Iso-1-Cytochrome C (T12A) : Yeast Cytochrome C Peroxidase Complex
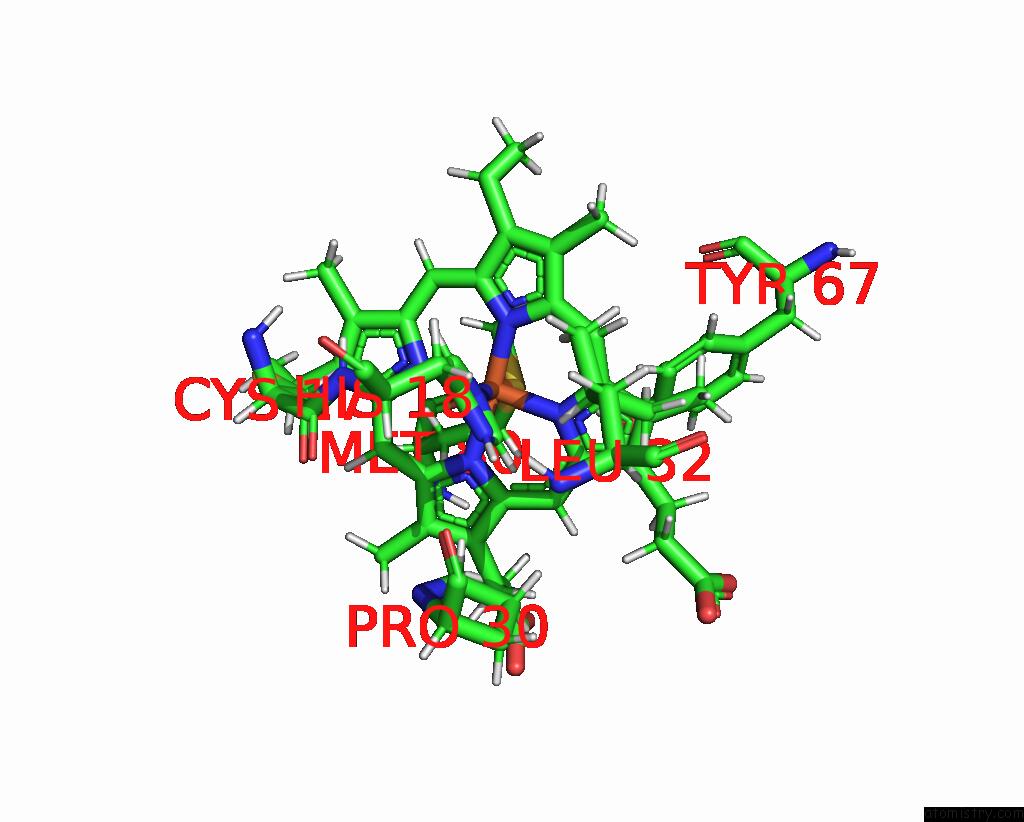
Mono view
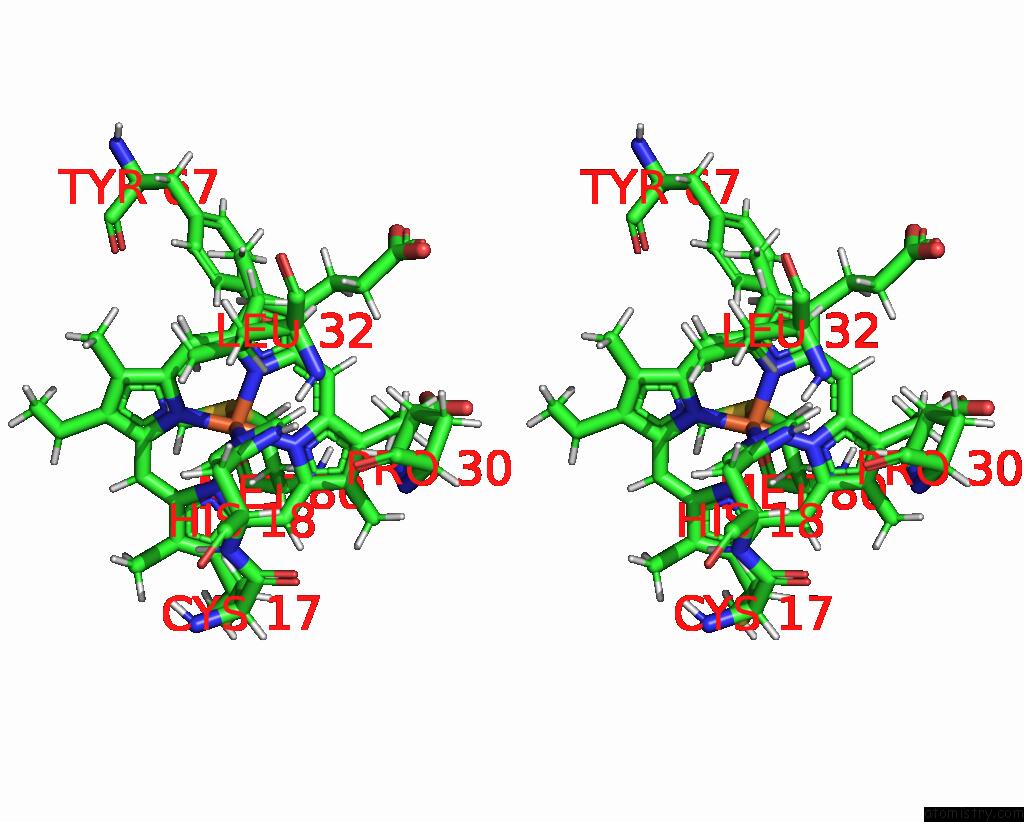
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Solution Structure of the Yeast Iso-1-Cytochrome C (T12A) : Yeast Cytochrome C Peroxidase Complex within 5.0Å range:
|
Reference:
A.N.Volkov,
Q.Bashir,
J.A.Worrall,
G.M.Ullmann,
M.Ubbink.
Shifting the Equilibrium Between the Encounter State and the Specific Form of A Protein Complex By Interfacial Point Mutations. J.Am.Chem.Soc. V. 132 11487 2010.
ISSN: ISSN 0002-7863
PubMed: 20672804
DOI: 10.1021/JA100867C
Page generated: Sat Aug 3 23:52:32 2024
ISSN: ISSN 0002-7863
PubMed: 20672804
DOI: 10.1021/JA100867C
Last articles
F in 6IC6F in 6IC2
F in 6I77
F in 6I78
F in 6I76
F in 6I74
F in 6I75
F in 6I4B
F in 6I4I
F in 6I2Y