Iron »
PDB 3b9j-3by0 »
3bnd »
Iron in PDB 3bnd: Lipoxygenase-1 (Soybean), I553V Mutant
Enzymatic activity of Lipoxygenase-1 (Soybean), I553V Mutant
All present enzymatic activity of Lipoxygenase-1 (Soybean), I553V Mutant:
1.13.11.12;
1.13.11.12;
Protein crystallography data
The structure of Lipoxygenase-1 (Soybean), I553V Mutant, PDB code: 3bnd
was solved by
D.R.Tomchick,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 27.37 / 1.60 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 94.012, 93.004, 49.351, 90.00, 90.61, 90.00 |
| R / Rfree (%) | 16.8 / 20 |
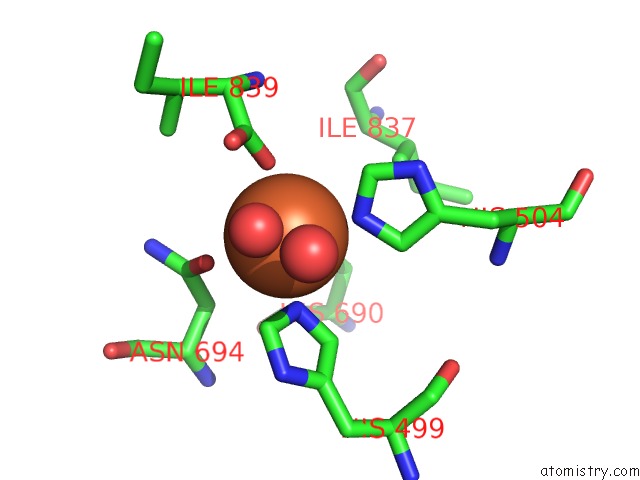
Iron Binding Sites:
The binding sites of Iron atom in the Lipoxygenase-1 (Soybean), I553V Mutant
(pdb code 3bnd). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total only one binding site of Iron was determined in the Lipoxygenase-1 (Soybean), I553V Mutant, PDB code: 3bnd:
In total only one binding site of Iron was determined in the Lipoxygenase-1 (Soybean), I553V Mutant, PDB code: 3bnd:
Iron binding site 1 out of 1 in 3bnd
Go back to
Iron binding site 1 out
of 1 in the Lipoxygenase-1 (Soybean), I553V Mutant
Mono view
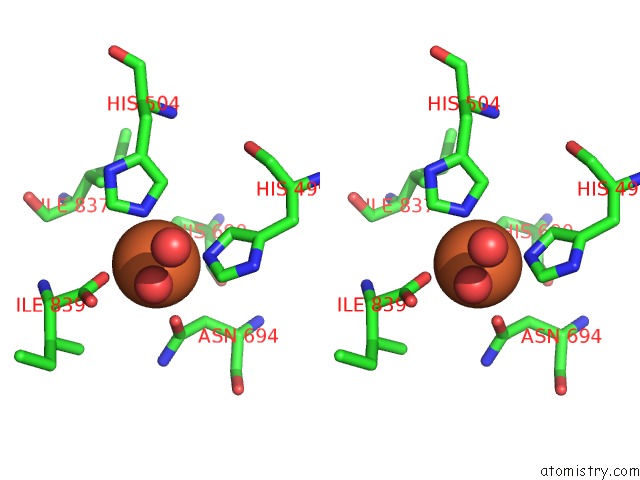
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Lipoxygenase-1 (Soybean), I553V Mutant within 5.0Å range:
|
Reference:
M.P.Meyer,
D.R.Tomchick,
J.P.Klinman.
Enzyme Structure and Dynamics Affect Hydrogen Tunneling: the Impact of A Remote Side Chain (I553) in Soybean Lipoxygenase-1. Proc.Natl.Acad.Sci.Usa V. 105 1146 2008.
ISSN: ISSN 0027-8424
PubMed: 18216254
DOI: 10.1073/PNAS.0710643105
Page generated: Mon Aug 4 23:57:47 2025
ISSN: ISSN 0027-8424
PubMed: 18216254
DOI: 10.1073/PNAS.0710643105
Last articles
Zn in 1JZQZn in 1JZS
Zn in 1JXP
Zn in 1JYB
Zn in 1JY8
Zn in 1JWH
Zn in 1JWQ
Zn in 1JV0
Zn in 1JWB
Zn in 1JW9