Iron »
PDB 3crv-3dby »
3cv8 »
Iron in PDB 3cv8: Crystal Structure of Vitamin D Hydroxylase Cytochrome P450 105A1 (R84F Mutant)
Enzymatic activity of Crystal Structure of Vitamin D Hydroxylase Cytochrome P450 105A1 (R84F Mutant)
All present enzymatic activity of Crystal Structure of Vitamin D Hydroxylase Cytochrome P450 105A1 (R84F Mutant):
1.14.14.1;
1.14.14.1;
Protein crystallography data
The structure of Crystal Structure of Vitamin D Hydroxylase Cytochrome P450 105A1 (R84F Mutant), PDB code: 3cv8
was solved by
K.Hayashi,
H.Sugimoto,
R.Shinkyo,
M.Yamada,
S.Ikeda,
S.Ikushiro,
M.Kamakura,
Y.Shiro,
T.Sakaki,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.00 / 2.00 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 52.500, 53.384, 138.970, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.1 / 23 |
Iron Binding Sites:
The binding sites of Iron atom in the Crystal Structure of Vitamin D Hydroxylase Cytochrome P450 105A1 (R84F Mutant)
(pdb code 3cv8). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total only one binding site of Iron was determined in the Crystal Structure of Vitamin D Hydroxylase Cytochrome P450 105A1 (R84F Mutant), PDB code: 3cv8:
In total only one binding site of Iron was determined in the Crystal Structure of Vitamin D Hydroxylase Cytochrome P450 105A1 (R84F Mutant), PDB code: 3cv8:
Iron binding site 1 out of 1 in 3cv8
Go back to
Iron binding site 1 out
of 1 in the Crystal Structure of Vitamin D Hydroxylase Cytochrome P450 105A1 (R84F Mutant)
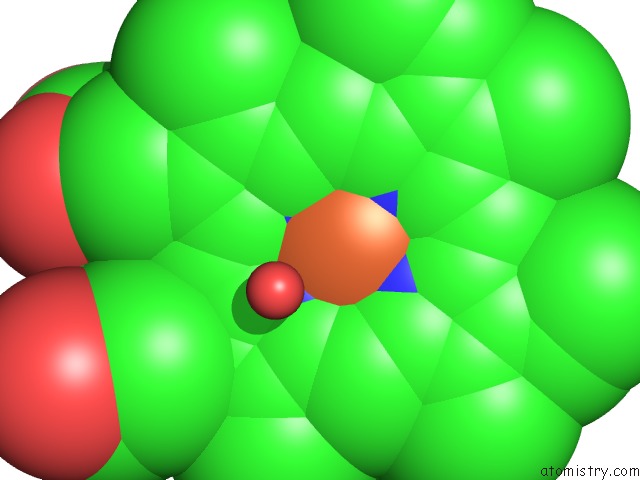
Mono view
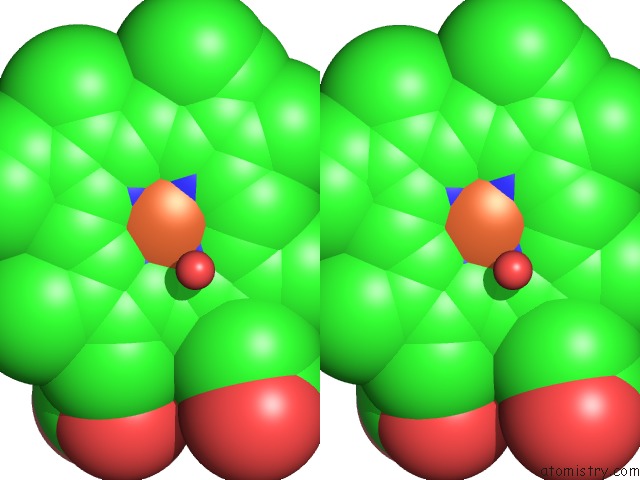
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Crystal Structure of Vitamin D Hydroxylase Cytochrome P450 105A1 (R84F Mutant) within 5.0Å range:
|
Reference:
K.Hayashi,
H.Sugimoto,
R.Shinkyo,
M.Yamada,
S.Ikeda,
S.Ikushiro,
M.Kamakura,
Y.Shiro,
T.Sakaki.
Structure-Based Design of A Highly Active Vitamin D Hydroxylase From Streptomyces Griseolus CYP105A1 Biochemistry V. 47 11964 2008.
ISSN: ISSN 0006-2960
PubMed: 18937506
DOI: 10.1021/BI801222D
Page generated: Tue Aug 5 00:18:19 2025
ISSN: ISSN 0006-2960
PubMed: 18937506
DOI: 10.1021/BI801222D
Last articles
Xe in 2OQUXe in 2IE6
Xe in 2IC0
Xe in 2DKI
Xe in 2FIC
Xe in 2A7A
Xe in 2A7D
Xe in 2A9R
Xe in 2A7B
Xe in 1ZDM