Iron »
PDB 3fpv-3gcj »
3g93 »
Iron in PDB 3g93: Single Ligand Occupancy Crystal Structure of Cytochrome P450 2B4 in Complex with the Inhibitor 1-Biphenyl-4-Methyl-1H-Imidazole
Enzymatic activity of Single Ligand Occupancy Crystal Structure of Cytochrome P450 2B4 in Complex with the Inhibitor 1-Biphenyl-4-Methyl-1H-Imidazole
All present enzymatic activity of Single Ligand Occupancy Crystal Structure of Cytochrome P450 2B4 in Complex with the Inhibitor 1-Biphenyl-4-Methyl-1H-Imidazole:
1.14.14.1;
1.14.14.1;
Protein crystallography data
The structure of Single Ligand Occupancy Crystal Structure of Cytochrome P450 2B4 in Complex with the Inhibitor 1-Biphenyl-4-Methyl-1H-Imidazole, PDB code: 3g93
was solved by
S.C.Gay,
L.Sun,
K.Maekawa,
J.R.Halpert,
C.D.Stout,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.75 / 3.20 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 86.910, 151.815, 181.583, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 23.7 / 27 |
Iron Binding Sites:
The binding sites of Iron atom in the Single Ligand Occupancy Crystal Structure of Cytochrome P450 2B4 in Complex with the Inhibitor 1-Biphenyl-4-Methyl-1H-Imidazole
(pdb code 3g93). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 4 binding sites of Iron where determined in the Single Ligand Occupancy Crystal Structure of Cytochrome P450 2B4 in Complex with the Inhibitor 1-Biphenyl-4-Methyl-1H-Imidazole, PDB code: 3g93:
Jump to Iron binding site number: 1; 2; 3; 4;
In total 4 binding sites of Iron where determined in the Single Ligand Occupancy Crystal Structure of Cytochrome P450 2B4 in Complex with the Inhibitor 1-Biphenyl-4-Methyl-1H-Imidazole, PDB code: 3g93:
Jump to Iron binding site number: 1; 2; 3; 4;
Iron binding site 1 out of 4 in 3g93
Go back to
Iron binding site 1 out
of 4 in the Single Ligand Occupancy Crystal Structure of Cytochrome P450 2B4 in Complex with the Inhibitor 1-Biphenyl-4-Methyl-1H-Imidazole
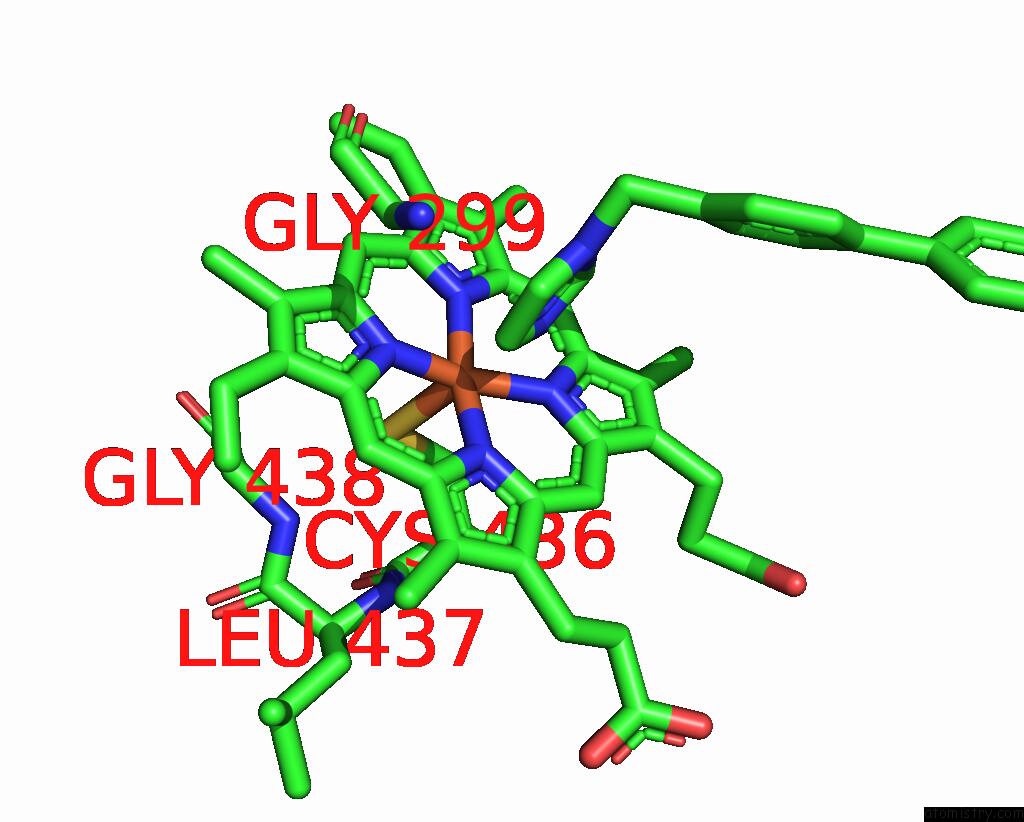
Mono view
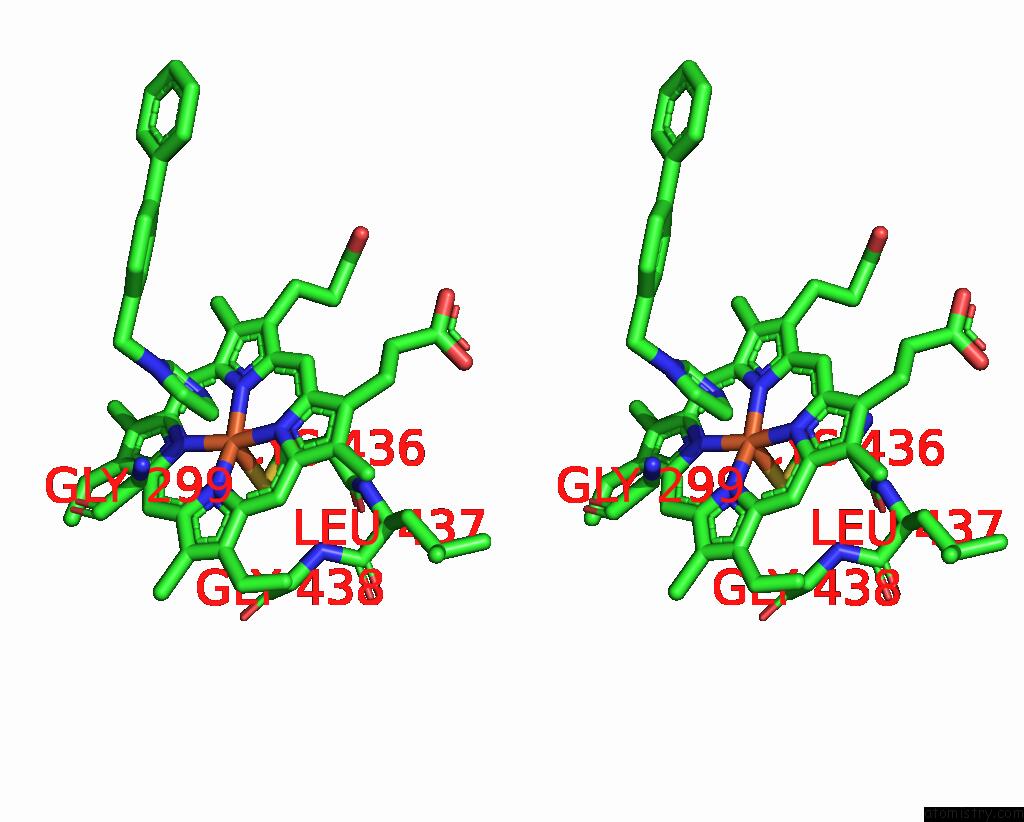
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Single Ligand Occupancy Crystal Structure of Cytochrome P450 2B4 in Complex with the Inhibitor 1-Biphenyl-4-Methyl-1H-Imidazole within 5.0Å range:
|
Iron binding site 2 out of 4 in 3g93
Go back to
Iron binding site 2 out
of 4 in the Single Ligand Occupancy Crystal Structure of Cytochrome P450 2B4 in Complex with the Inhibitor 1-Biphenyl-4-Methyl-1H-Imidazole
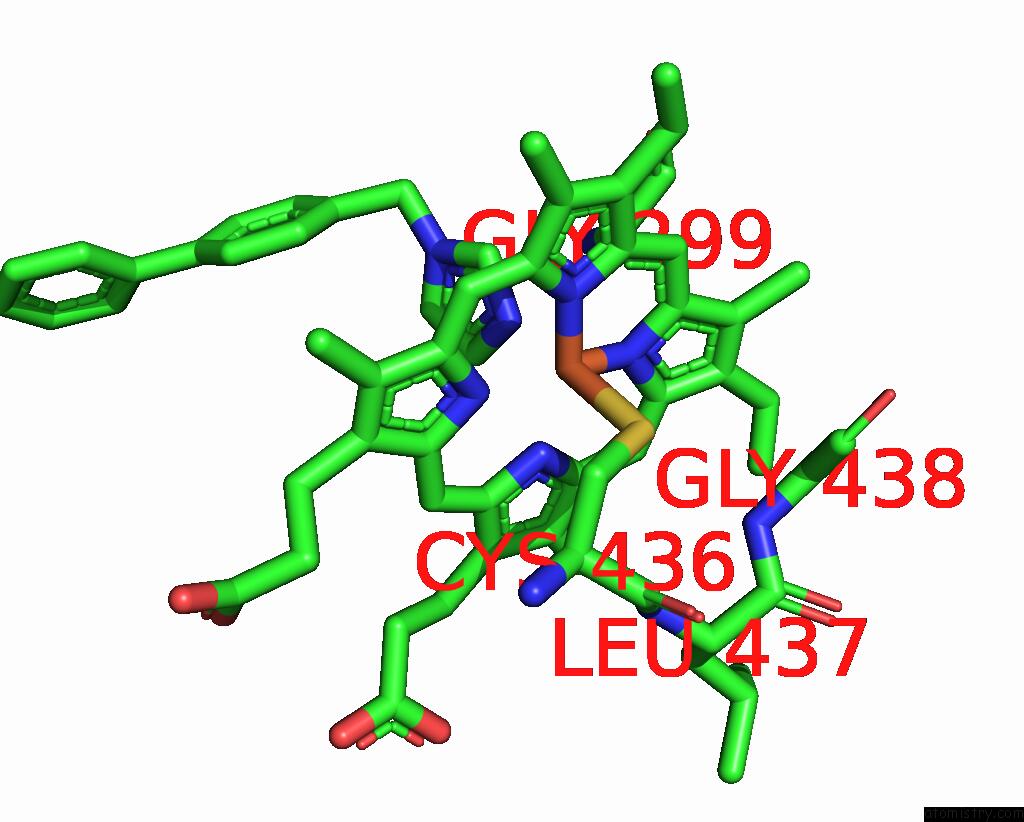
Mono view
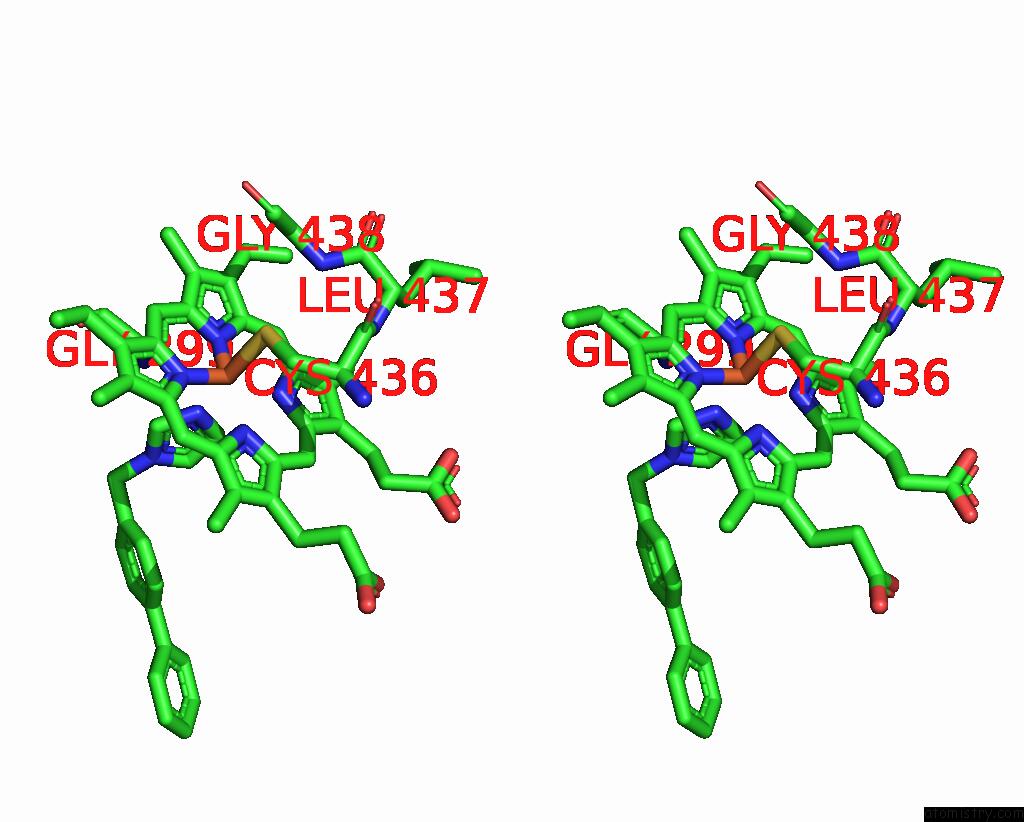
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Single Ligand Occupancy Crystal Structure of Cytochrome P450 2B4 in Complex with the Inhibitor 1-Biphenyl-4-Methyl-1H-Imidazole within 5.0Å range:
|
Iron binding site 3 out of 4 in 3g93
Go back to
Iron binding site 3 out
of 4 in the Single Ligand Occupancy Crystal Structure of Cytochrome P450 2B4 in Complex with the Inhibitor 1-Biphenyl-4-Methyl-1H-Imidazole
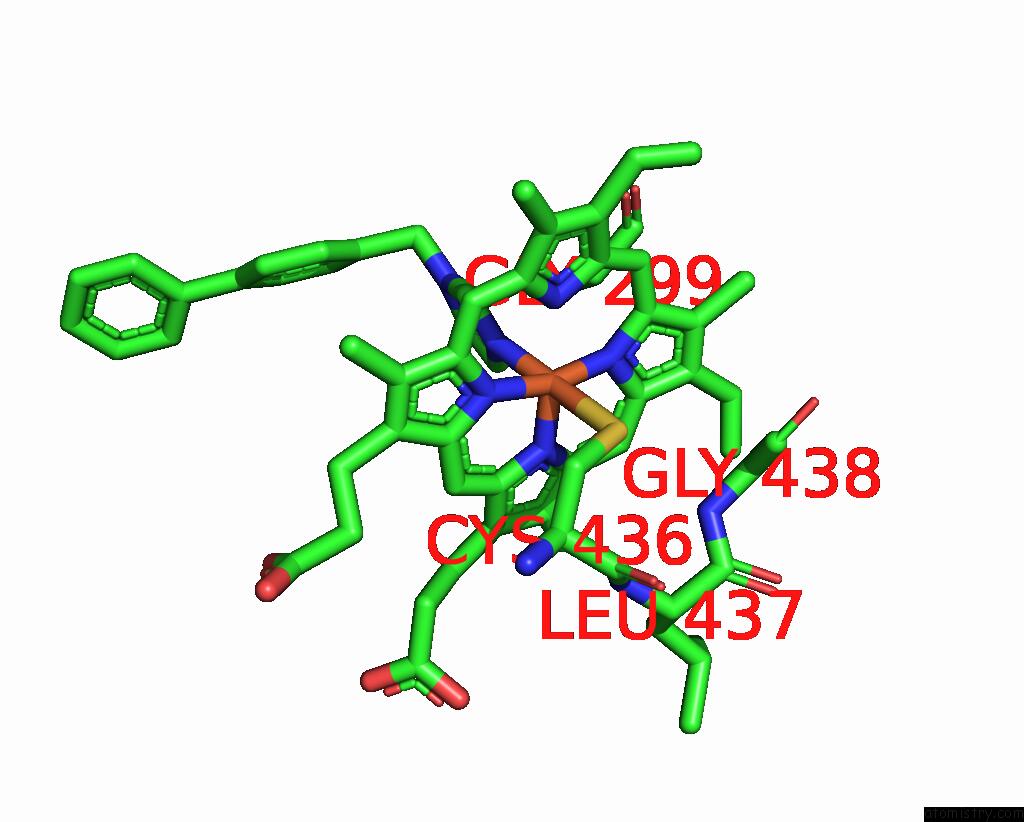
Mono view
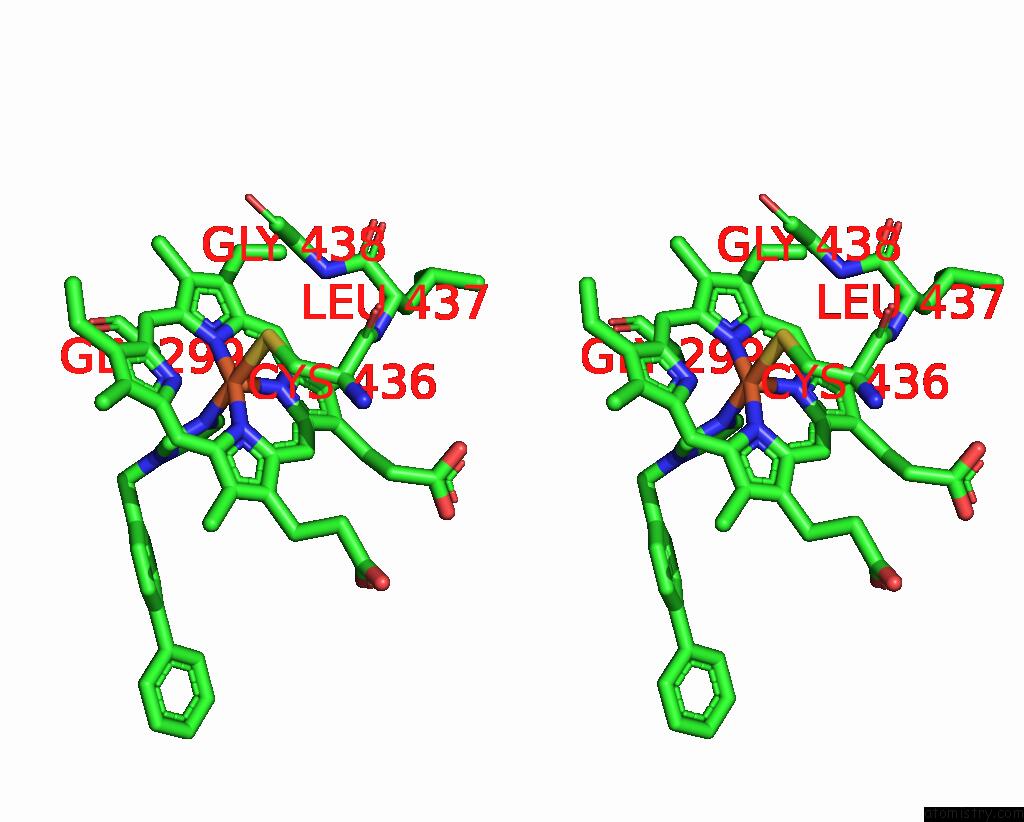
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 3 of Single Ligand Occupancy Crystal Structure of Cytochrome P450 2B4 in Complex with the Inhibitor 1-Biphenyl-4-Methyl-1H-Imidazole within 5.0Å range:
|
Iron binding site 4 out of 4 in 3g93
Go back to
Iron binding site 4 out
of 4 in the Single Ligand Occupancy Crystal Structure of Cytochrome P450 2B4 in Complex with the Inhibitor 1-Biphenyl-4-Methyl-1H-Imidazole
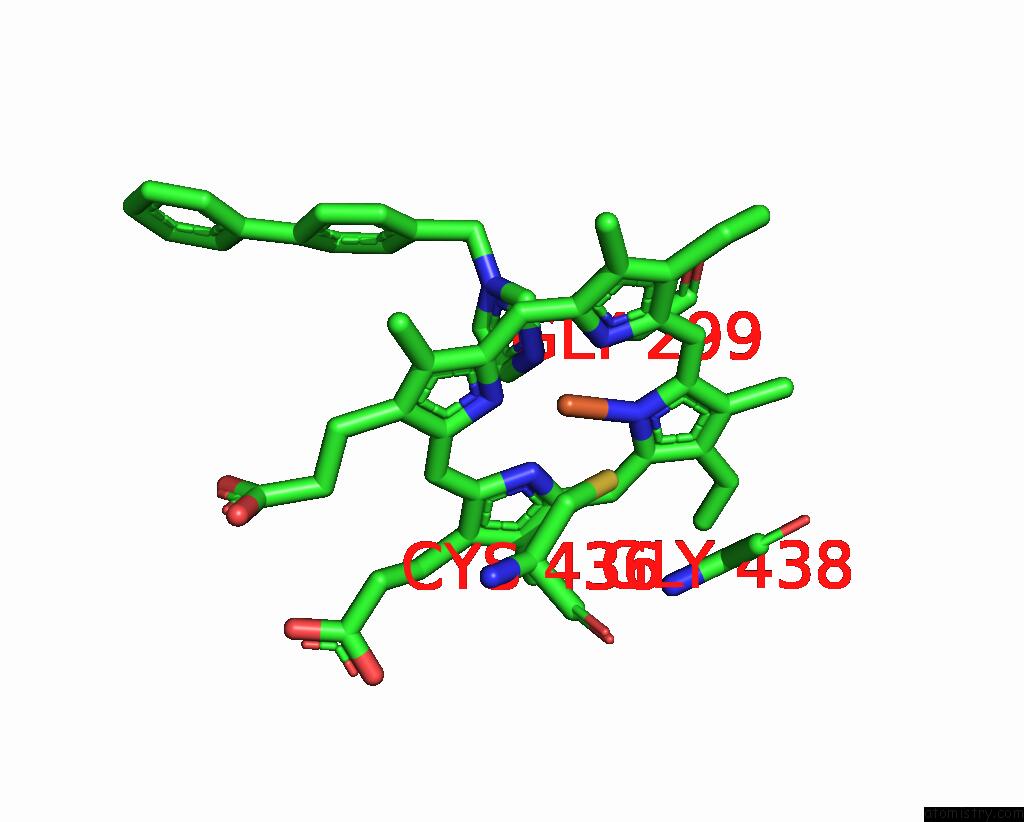
Mono view
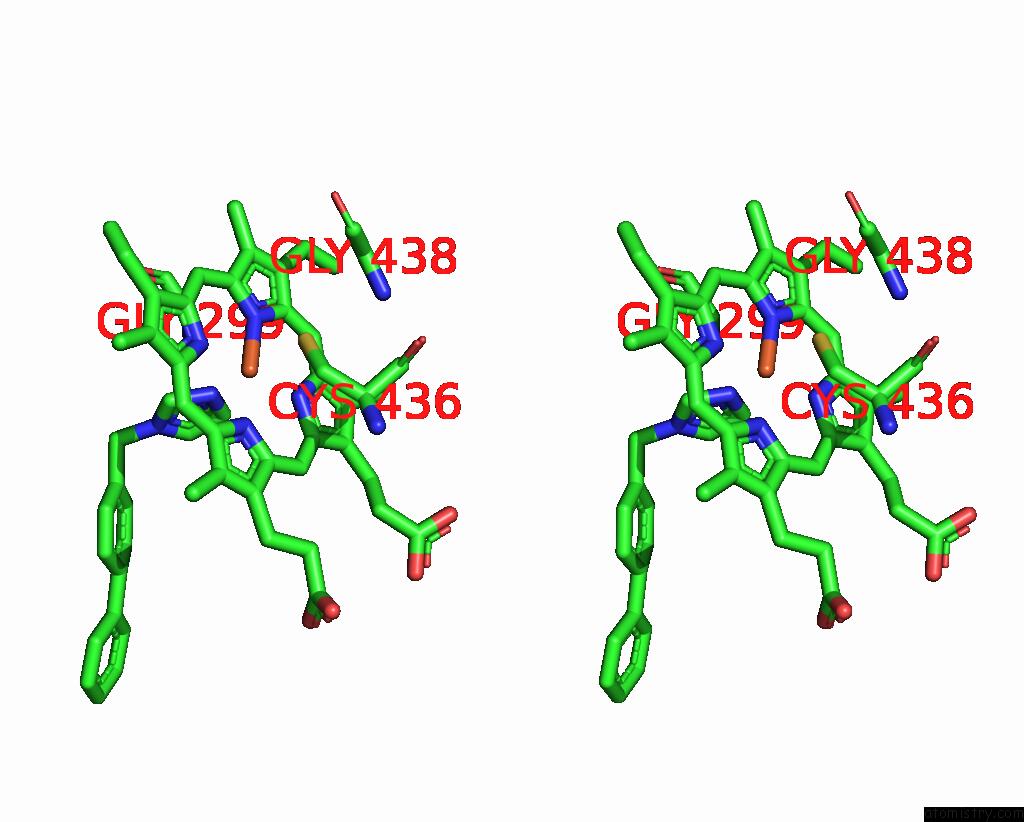
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 4 of Single Ligand Occupancy Crystal Structure of Cytochrome P450 2B4 in Complex with the Inhibitor 1-Biphenyl-4-Methyl-1H-Imidazole within 5.0Å range:
|
Reference:
S.C.Gay,
L.Sun,
K.Maekawa,
J.R.Halpert,
C.D.Stout.
Crystal Structures of Cytochrome P450 2B4 in Complex with the Inhibitor 1-Biphenyl-4-Methyl-1H-Imidazole: Ligand-Induced Structural Response Through Alpha-Helical Repositioning. Biochemistry V. 48 4762 2009.
ISSN: ISSN 0006-2960
PubMed: 19397311
DOI: 10.1021/BI9003765
Page generated: Tue Aug 5 01:22:55 2025
ISSN: ISSN 0006-2960
PubMed: 19397311
DOI: 10.1021/BI9003765
Last articles
Na in 7MD1Na in 7MD0
Na in 7MCY
Na in 7MCP
Na in 7MCQ
Na in 7MCU
Na in 7MCT
Na in 7MCN
Na in 7MCB
Na in 7MCL