Iron »
PDB 3jbt-3k9y »
3jx4 »
Iron in PDB 3jx4: Structure of Rat Neuronal Nitric Oxide Synthase D597N/M336V Mutant Heme Domain in Complex with N1-{(3'R,4'S)-4'-[(6"-Amino-4"- Methylpyridin-2"-Yl)Methyl]Pyrrolidin-3'-Yl}-N2-(3'-Fluorophenethyl) Ethane-1,2-Diamine
Enzymatic activity of Structure of Rat Neuronal Nitric Oxide Synthase D597N/M336V Mutant Heme Domain in Complex with N1-{(3'R,4'S)-4'-[(6"-Amino-4"- Methylpyridin-2"-Yl)Methyl]Pyrrolidin-3'-Yl}-N2-(3'-Fluorophenethyl) Ethane-1,2-Diamine
All present enzymatic activity of Structure of Rat Neuronal Nitric Oxide Synthase D597N/M336V Mutant Heme Domain in Complex with N1-{(3'R,4'S)-4'-[(6"-Amino-4"- Methylpyridin-2"-Yl)Methyl]Pyrrolidin-3'-Yl}-N2-(3'-Fluorophenethyl) Ethane-1,2-Diamine:
1.14.13.39;
1.14.13.39;
Protein crystallography data
The structure of Structure of Rat Neuronal Nitric Oxide Synthase D597N/M336V Mutant Heme Domain in Complex with N1-{(3'R,4'S)-4'-[(6"-Amino-4"- Methylpyridin-2"-Yl)Methyl]Pyrrolidin-3'-Yl}-N2-(3'-Fluorophenethyl) Ethane-1,2-Diamine, PDB code: 3jx4
was solved by
S.L.Delker,
H.Li,
T.L.Poulos,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 49.89 / 2.26 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 51.602, 110.975, 164.866, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 21.1 / 26.6 |
Other elements in 3jx4:
The structure of Structure of Rat Neuronal Nitric Oxide Synthase D597N/M336V Mutant Heme Domain in Complex with N1-{(3'R,4'S)-4'-[(6"-Amino-4"- Methylpyridin-2"-Yl)Methyl]Pyrrolidin-3'-Yl}-N2-(3'-Fluorophenethyl) Ethane-1,2-Diamine also contains other interesting chemical elements:
| Fluorine | (F) | 2 atoms |
| Zinc | (Zn) | 1 atom |
Iron Binding Sites:
The binding sites of Iron atom in the Structure of Rat Neuronal Nitric Oxide Synthase D597N/M336V Mutant Heme Domain in Complex with N1-{(3'R,4'S)-4'-[(6"-Amino-4"- Methylpyridin-2"-Yl)Methyl]Pyrrolidin-3'-Yl}-N2-(3'-Fluorophenethyl) Ethane-1,2-Diamine
(pdb code 3jx4). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 2 binding sites of Iron where determined in the Structure of Rat Neuronal Nitric Oxide Synthase D597N/M336V Mutant Heme Domain in Complex with N1-{(3'R,4'S)-4'-[(6"-Amino-4"- Methylpyridin-2"-Yl)Methyl]Pyrrolidin-3'-Yl}-N2-(3'-Fluorophenethyl) Ethane-1,2-Diamine, PDB code: 3jx4:
Jump to Iron binding site number: 1; 2;
In total 2 binding sites of Iron where determined in the Structure of Rat Neuronal Nitric Oxide Synthase D597N/M336V Mutant Heme Domain in Complex with N1-{(3'R,4'S)-4'-[(6"-Amino-4"- Methylpyridin-2"-Yl)Methyl]Pyrrolidin-3'-Yl}-N2-(3'-Fluorophenethyl) Ethane-1,2-Diamine, PDB code: 3jx4:
Jump to Iron binding site number: 1; 2;
Iron binding site 1 out of 2 in 3jx4
Go back to
Iron binding site 1 out
of 2 in the Structure of Rat Neuronal Nitric Oxide Synthase D597N/M336V Mutant Heme Domain in Complex with N1-{(3'R,4'S)-4'-[(6"-Amino-4"- Methylpyridin-2"-Yl)Methyl]Pyrrolidin-3'-Yl}-N2-(3'-Fluorophenethyl) Ethane-1,2-Diamine
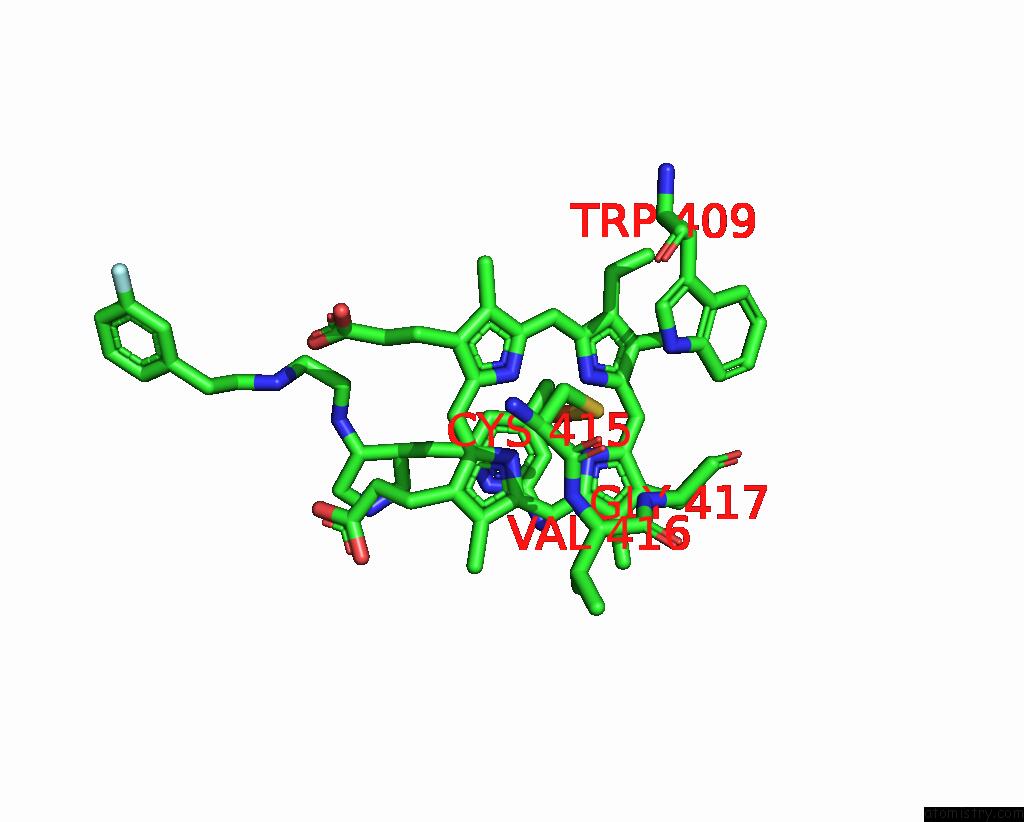
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Structure of Rat Neuronal Nitric Oxide Synthase D597N/M336V Mutant Heme Domain in Complex with N1-{(3'R,4'S)-4'-[(6"-Amino-4"- Methylpyridin-2"-Yl)Methyl]Pyrrolidin-3'-Yl}-N2-(3'-Fluorophenethyl) Ethane-1,2-Diamine within 5.0Å range:
|
Iron binding site 2 out of 2 in 3jx4
Go back to
Iron binding site 2 out
of 2 in the Structure of Rat Neuronal Nitric Oxide Synthase D597N/M336V Mutant Heme Domain in Complex with N1-{(3'R,4'S)-4'-[(6"-Amino-4"- Methylpyridin-2"-Yl)Methyl]Pyrrolidin-3'-Yl}-N2-(3'-Fluorophenethyl) Ethane-1,2-Diamine
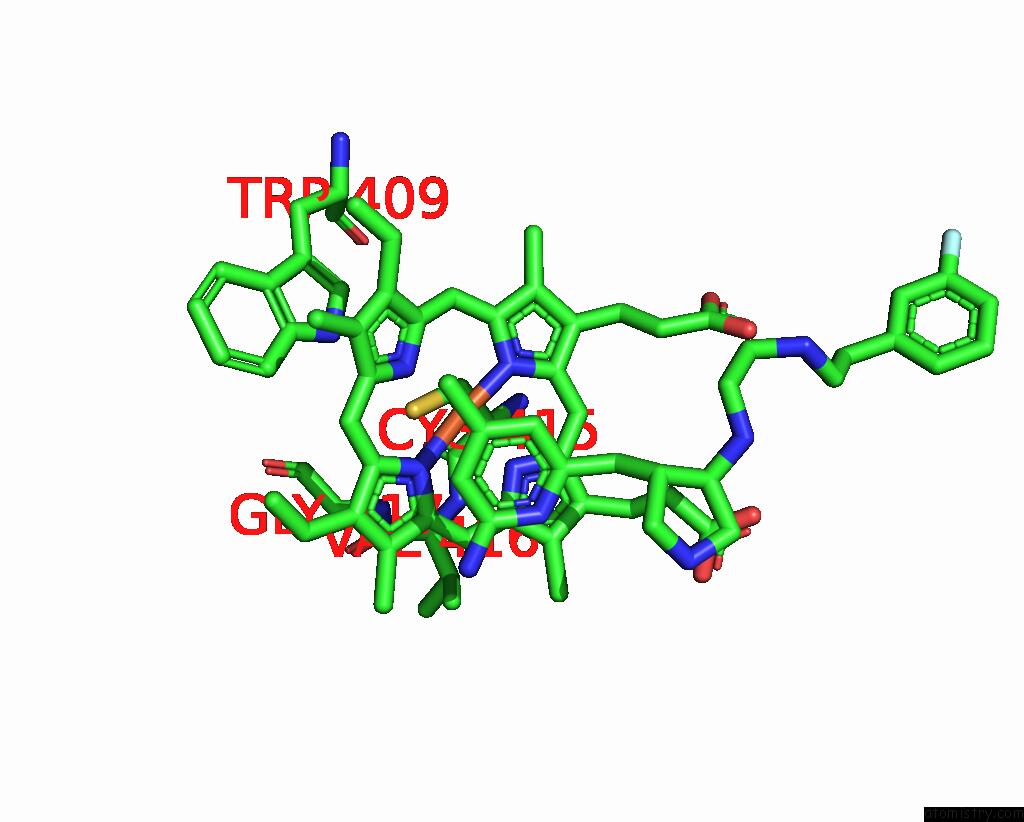
Mono view
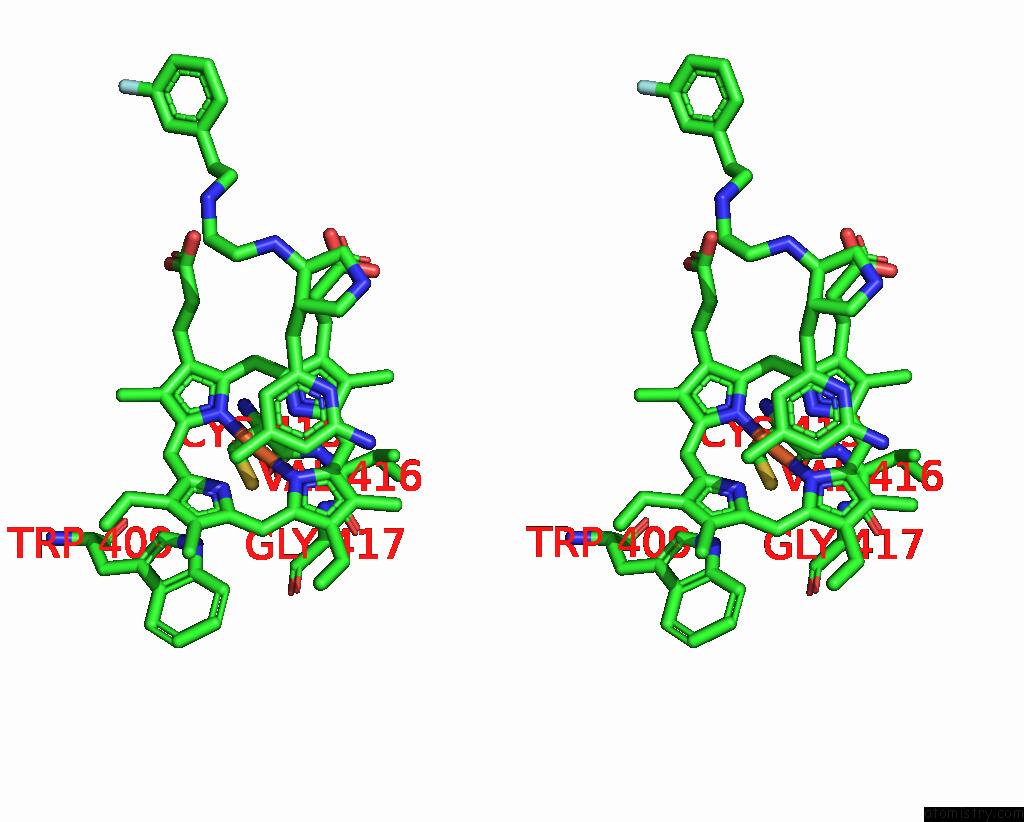
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Structure of Rat Neuronal Nitric Oxide Synthase D597N/M336V Mutant Heme Domain in Complex with N1-{(3'R,4'S)-4'-[(6"-Amino-4"- Methylpyridin-2"-Yl)Methyl]Pyrrolidin-3'-Yl}-N2-(3'-Fluorophenethyl) Ethane-1,2-Diamine within 5.0Å range:
|
Reference:
S.L.Delker,
H.Ji,
H.Li,
J.Jamal,
J.Fang,
F.Xue,
R.B.Silverman,
T.L.Poulos.
Unexpected Binding Modes of Nitric Oxide Synthase Inhibitors Effective in the Prevention of A Cerebral Palsy Phenotype in An Animal Model. J.Am.Chem.Soc. V. 132 5437 2010.
ISSN: ISSN 0002-7863
PubMed: 20337441
DOI: 10.1021/JA910228A
Page generated: Tue Aug 5 03:08:00 2025
ISSN: ISSN 0002-7863
PubMed: 20337441
DOI: 10.1021/JA910228A
Last articles
K in 5COGK in 5CP4
K in 5CVT
K in 5CO0
K in 5CK6
K in 5CMX
K in 5CK0
K in 5CJZ
K in 5CC8
K in 5CDB