Iron »
PDB 4cuo-4d36 »
4d02 »
Iron in PDB 4d02: The Crystallographic Structure of Flavorubredoxin From Escherichia Coli
Protein crystallography data
The structure of The Crystallographic Structure of Flavorubredoxin From Escherichia Coli, PDB code: 4d02
was solved by
C.V.Romao,
J.B.Vicente,
T.Bandeiras,
M.A.Carrondo,
M.Teixeira,
C.Frazao,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 129.496 / 1.76 |
| Space group | P 6 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 149.529, 149.529, 94.498, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 14.07 / n/a |
Other elements in 4d02:
The structure of The Crystallographic Structure of Flavorubredoxin From Escherichia Coli also contains other interesting chemical elements:
| Chlorine | (Cl) | 2 atoms |
Iron Binding Sites:
The binding sites of Iron atom in the The Crystallographic Structure of Flavorubredoxin From Escherichia Coli
(pdb code 4d02). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 2 binding sites of Iron where determined in the The Crystallographic Structure of Flavorubredoxin From Escherichia Coli, PDB code: 4d02:
Jump to Iron binding site number: 1; 2;
In total 2 binding sites of Iron where determined in the The Crystallographic Structure of Flavorubredoxin From Escherichia Coli, PDB code: 4d02:
Jump to Iron binding site number: 1; 2;
Iron binding site 1 out of 2 in 4d02
Go back to
Iron binding site 1 out
of 2 in the The Crystallographic Structure of Flavorubredoxin From Escherichia Coli
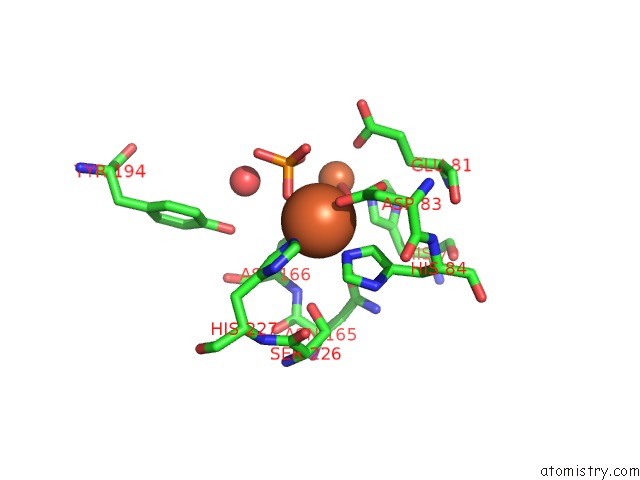
Mono view
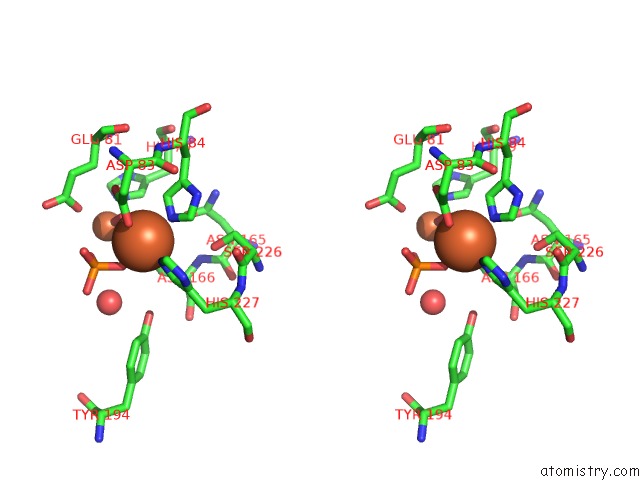
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of The Crystallographic Structure of Flavorubredoxin From Escherichia Coli within 5.0Å range:
|
Iron binding site 2 out of 2 in 4d02
Go back to
Iron binding site 2 out
of 2 in the The Crystallographic Structure of Flavorubredoxin From Escherichia Coli
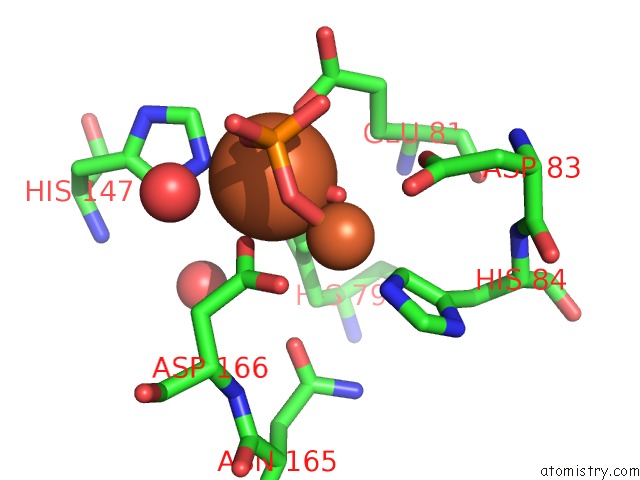
Mono view
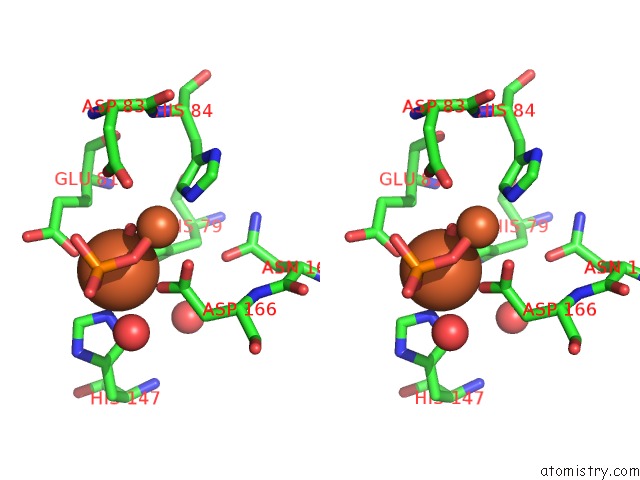
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of The Crystallographic Structure of Flavorubredoxin From Escherichia Coli within 5.0Å range:
|
Reference:
V.L.Goncalves,
J.B.Vicente,
L.Pinto,
C.V.Romao,
C.Frazao,
P.Sarti,
A.Giuffre,
M.Teixeira.
Flavodiiron Oxygen Reductase From Entamoeba Histolytica: Modulation of Substrate Preference By Tyrosine 271 and Lysine 53. J.Biol.Chem. V. 289 28260 2014.
ISSN: ISSN 0021-9258
PubMed: 25151360
DOI: 10.1074/JBC.M114.579086
Page generated: Tue Aug 5 09:41:28 2025
ISSN: ISSN 0021-9258
PubMed: 25151360
DOI: 10.1074/JBC.M114.579086
Last articles
Mg in 2OQYMg in 2OUP
Mg in 2OUN
Mg in 2OU7
Mg in 2OTG
Mg in 2OSB
Mg in 2OS8
Mg in 2ORI
Mg in 2ORW
Mg in 2ONP