Iron »
PDB 5mms-5o10 »
5nku »
Iron in PDB 5nku: Joint Neutron/X-Ray Structure of Dimeric Chlorite Dismutase From Cyanothece Sp. PCC7425
Protein crystallography data
The structure of Joint Neutron/X-Ray Structure of Dimeric Chlorite Dismutase From Cyanothece Sp. PCC7425, PDB code: 5nku
was solved by
D.Puehringer,
I.Schaffner,
G.Mlynek,
C.Obinger,
K.Djinovic-Carugo,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | N/A / 2.00 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 52.427, 53.015, 55.337, 107.30, 98.54, 109.86 |
| R / Rfree (%) | 23.9 / 25.3 |
Other elements in 5nku:
The structure of Joint Neutron/X-Ray Structure of Dimeric Chlorite Dismutase From Cyanothece Sp. PCC7425 also contains other interesting chemical elements:
| Chlorine | (Cl) | 1 atom |
Iron Binding Sites:
The binding sites of Iron atom in the Joint Neutron/X-Ray Structure of Dimeric Chlorite Dismutase From Cyanothece Sp. PCC7425
(pdb code 5nku). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 2 binding sites of Iron where determined in the Joint Neutron/X-Ray Structure of Dimeric Chlorite Dismutase From Cyanothece Sp. PCC7425, PDB code: 5nku:
Jump to Iron binding site number: 1; 2;
In total 2 binding sites of Iron where determined in the Joint Neutron/X-Ray Structure of Dimeric Chlorite Dismutase From Cyanothece Sp. PCC7425, PDB code: 5nku:
Jump to Iron binding site number: 1; 2;
Iron binding site 1 out of 2 in 5nku
Go back to
Iron binding site 1 out
of 2 in the Joint Neutron/X-Ray Structure of Dimeric Chlorite Dismutase From Cyanothece Sp. PCC7425
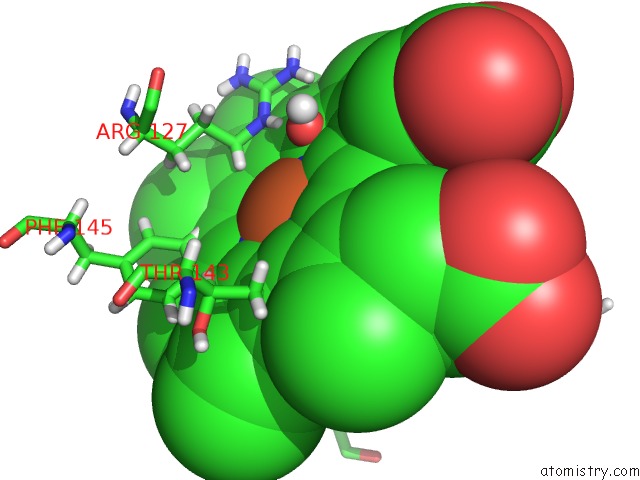
Mono view
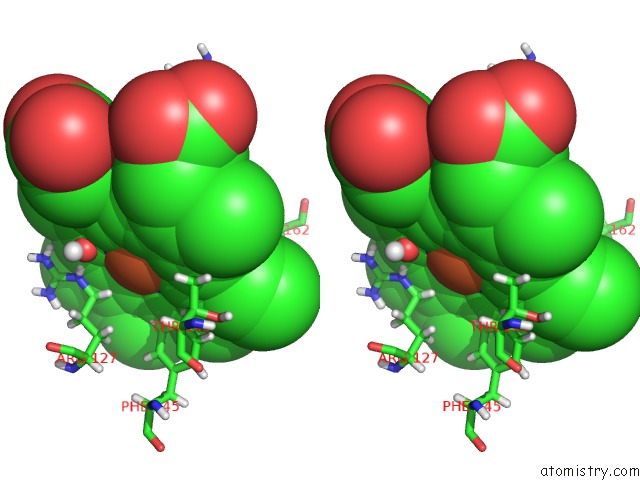
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Joint Neutron/X-Ray Structure of Dimeric Chlorite Dismutase From Cyanothece Sp. PCC7425 within 5.0Å range:
|
Iron binding site 2 out of 2 in 5nku
Go back to
Iron binding site 2 out
of 2 in the Joint Neutron/X-Ray Structure of Dimeric Chlorite Dismutase From Cyanothece Sp. PCC7425
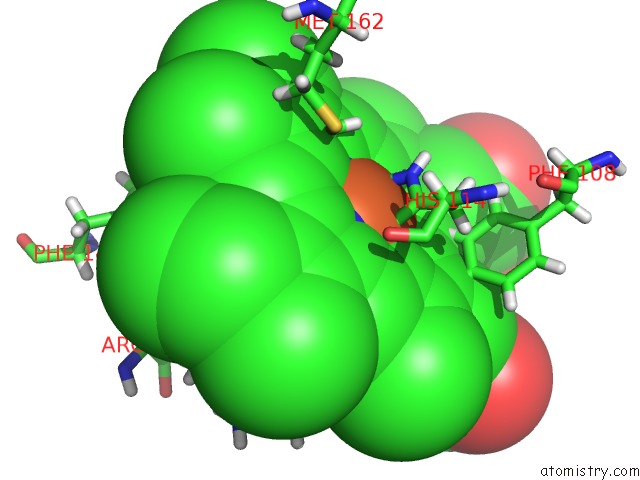
Mono view
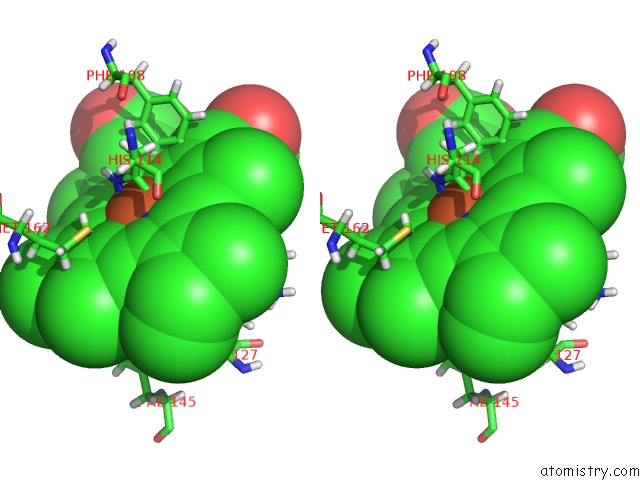
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Joint Neutron/X-Ray Structure of Dimeric Chlorite Dismutase From Cyanothece Sp. PCC7425 within 5.0Å range:
|
Reference:
I.Schaffner,
G.Mlynek,
N.Flego,
D.Puhringer,
J.Libiseller-Egger,
L.Coates,
S.Hofbauer,
M.Bellei,
P.G.Furtmuller,
G.Battistuzzi,
G.Smulevich,
K.Djinovic-Carugo,
C.Obinger.
Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights From Structural and Kinetic Studies. Acs Catal V. 7 7962 2017.
ISSN: ESSN 2155-5435
PubMed: 29142780
DOI: 10.1021/ACSCATAL.7B01749
Page generated: Wed Aug 6 00:06:19 2025
ISSN: ESSN 2155-5435
PubMed: 29142780
DOI: 10.1021/ACSCATAL.7B01749
Last articles
Mg in 1VQ8Mg in 1VQ4
Mg in 1VPA
Mg in 1VPE
Mg in 1VOM
Mg in 1VMA
Mg in 1VMK
Mg in 1VM9
Mg in 1VCR
Mg in 1VLB