Iron »
PDB 6tny-6u97 »
6u7t »
Iron in PDB 6u7t: Muty Adenine Glycosylase Bound to Dna Containing A Transition State Analog (1N) Paired with D(8-Oxo-G)
Enzymatic activity of Muty Adenine Glycosylase Bound to Dna Containing A Transition State Analog (1N) Paired with D(8-Oxo-G)
All present enzymatic activity of Muty Adenine Glycosylase Bound to Dna Containing A Transition State Analog (1N) Paired with D(8-Oxo-G):
3.2.2.31;
3.2.2.31;
Protein crystallography data
The structure of Muty Adenine Glycosylase Bound to Dna Containing A Transition State Analog (1N) Paired with D(8-Oxo-G), PDB code: 6u7t
was solved by
V.L.O'shea Murray,
S.Cao,
M.P.Horvath,
S.S.David,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 41.33 / 2.00 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 37.800, 86.490, 140.580, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 26.1 / 27 |
Other elements in 6u7t:
The structure of Muty Adenine Glycosylase Bound to Dna Containing A Transition State Analog (1N) Paired with D(8-Oxo-G) also contains other interesting chemical elements:
| Calcium | (Ca) | 2 atoms |
Iron Binding Sites:
The binding sites of Iron atom in the Muty Adenine Glycosylase Bound to Dna Containing A Transition State Analog (1N) Paired with D(8-Oxo-G)
(pdb code 6u7t). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 4 binding sites of Iron where determined in the Muty Adenine Glycosylase Bound to Dna Containing A Transition State Analog (1N) Paired with D(8-Oxo-G), PDB code: 6u7t:
Jump to Iron binding site number: 1; 2; 3; 4;
In total 4 binding sites of Iron where determined in the Muty Adenine Glycosylase Bound to Dna Containing A Transition State Analog (1N) Paired with D(8-Oxo-G), PDB code: 6u7t:
Jump to Iron binding site number: 1; 2; 3; 4;
Iron binding site 1 out of 4 in 6u7t
Go back to
Iron binding site 1 out
of 4 in the Muty Adenine Glycosylase Bound to Dna Containing A Transition State Analog (1N) Paired with D(8-Oxo-G)
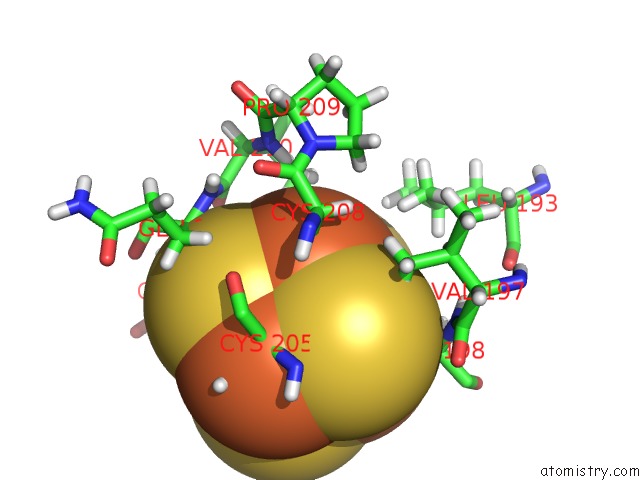
Mono view
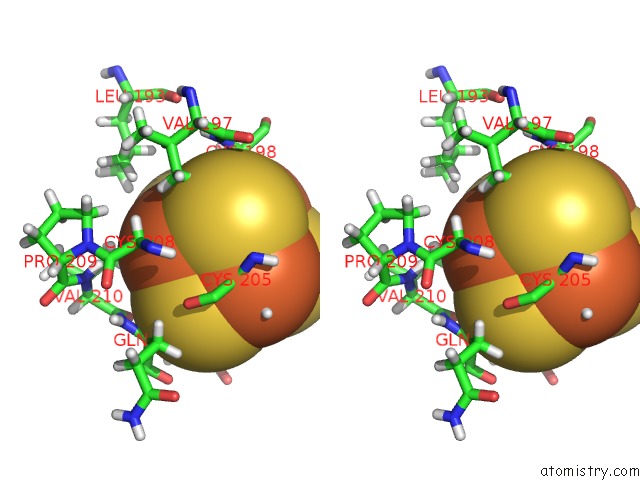
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Muty Adenine Glycosylase Bound to Dna Containing A Transition State Analog (1N) Paired with D(8-Oxo-G) within 5.0Å range:
|
Iron binding site 2 out of 4 in 6u7t
Go back to
Iron binding site 2 out
of 4 in the Muty Adenine Glycosylase Bound to Dna Containing A Transition State Analog (1N) Paired with D(8-Oxo-G)
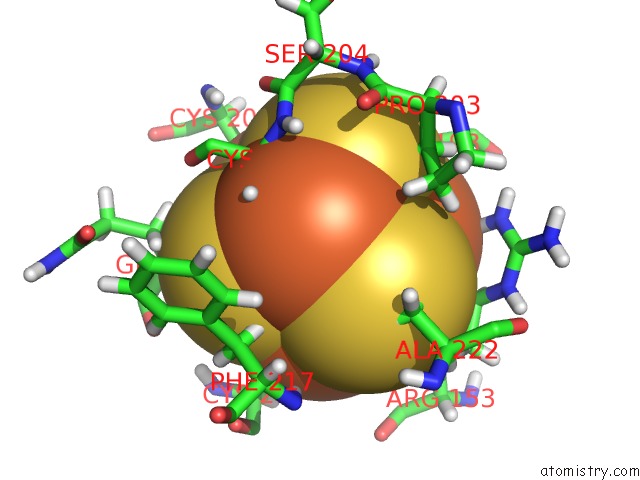
Mono view
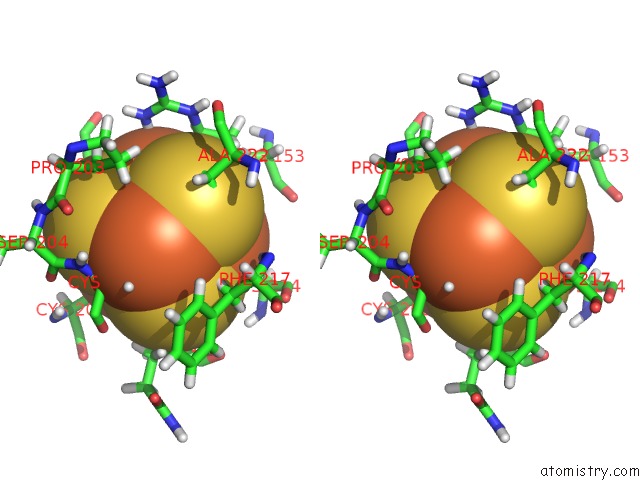
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Muty Adenine Glycosylase Bound to Dna Containing A Transition State Analog (1N) Paired with D(8-Oxo-G) within 5.0Å range:
|
Iron binding site 3 out of 4 in 6u7t
Go back to
Iron binding site 3 out
of 4 in the Muty Adenine Glycosylase Bound to Dna Containing A Transition State Analog (1N) Paired with D(8-Oxo-G)
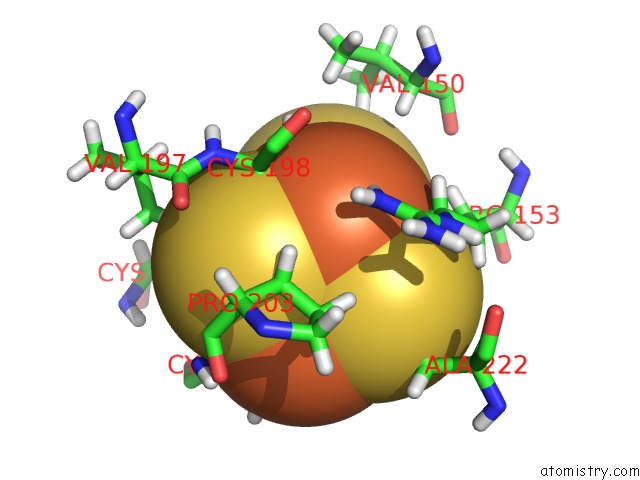
Mono view
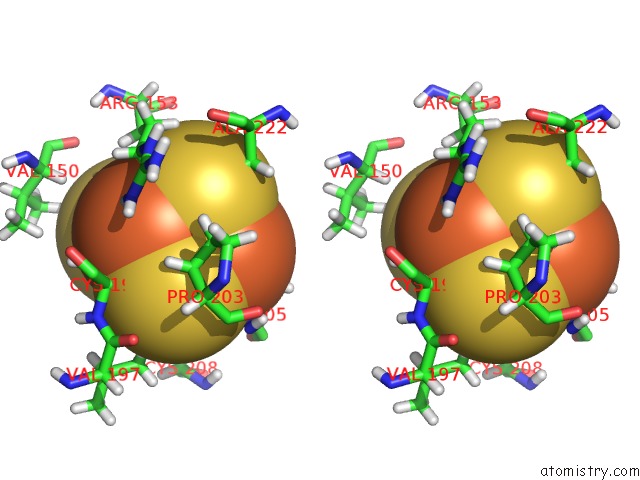
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 3 of Muty Adenine Glycosylase Bound to Dna Containing A Transition State Analog (1N) Paired with D(8-Oxo-G) within 5.0Å range:
|
Iron binding site 4 out of 4 in 6u7t
Go back to
Iron binding site 4 out
of 4 in the Muty Adenine Glycosylase Bound to Dna Containing A Transition State Analog (1N) Paired with D(8-Oxo-G)
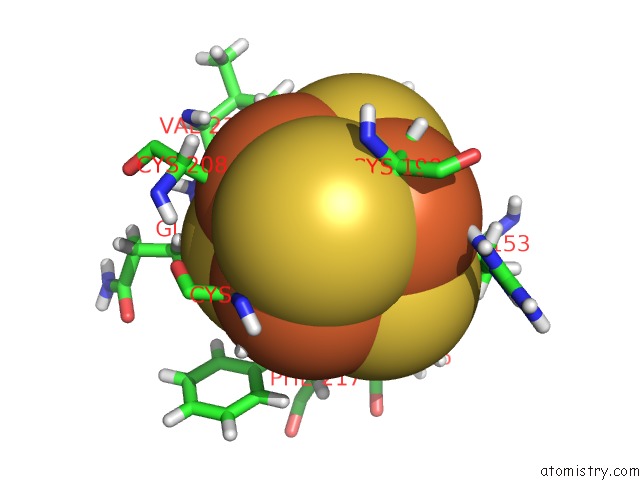
Mono view
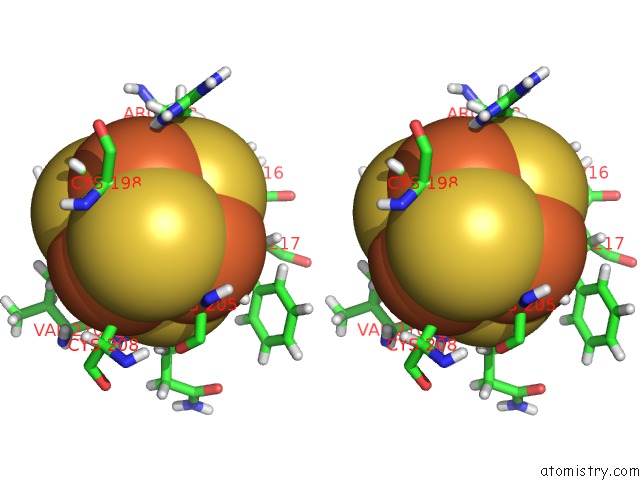
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 4 of Muty Adenine Glycosylase Bound to Dna Containing A Transition State Analog (1N) Paired with D(8-Oxo-G) within 5.0Å range:
|
Reference:
R.D.Woods,
V.L.O'shea,
A.Chu,
S.Cao,
J.L.Richards,
M.P.Horvath,
S.S.David.
Structure and Stereochemistry of the Base Excision Repair Glycosylase Muty Reveal A Mechanism Similar to Retaining Glycosidases. Nucleic Acids Res. V. 44 801 2016.
ISSN: ESSN 1362-4962
PubMed: 26673696
DOI: 10.1093/NAR/GKV1469
Page generated: Wed Aug 6 14:32:18 2025
ISSN: ESSN 1362-4962
PubMed: 26673696
DOI: 10.1093/NAR/GKV1469
Last articles
K in 7Q0GK in 7Q1C
K in 7Q1B
K in 7PXH
K in 7PZJ
K in 7PXG
K in 7PWY
K in 7PXE
K in 7PXF
K in 7PVC