Iron »
PDB 7lw8-7mh9 »
7m2u »
Iron in PDB 7m2u: Nucleotide Excision Repair Complex Tfiih RAD4-33
Enzymatic activity of Nucleotide Excision Repair Complex Tfiih RAD4-33
All present enzymatic activity of Nucleotide Excision Repair Complex Tfiih RAD4-33:
3.6.4.12;
3.6.4.12;
Other elements in 7m2u:
The structure of Nucleotide Excision Repair Complex Tfiih RAD4-33 also contains other interesting chemical elements:
| Calcium | (Ca) | 2 atoms |
| Zinc | (Zn) | 5 atoms |
Iron Binding Sites:
The binding sites of Iron atom in the Nucleotide Excision Repair Complex Tfiih RAD4-33
(pdb code 7m2u). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 4 binding sites of Iron where determined in the Nucleotide Excision Repair Complex Tfiih RAD4-33, PDB code: 7m2u:
Jump to Iron binding site number: 1; 2; 3; 4;
In total 4 binding sites of Iron where determined in the Nucleotide Excision Repair Complex Tfiih RAD4-33, PDB code: 7m2u:
Jump to Iron binding site number: 1; 2; 3; 4;
Iron binding site 1 out of 4 in 7m2u
Go back to
Iron binding site 1 out
of 4 in the Nucleotide Excision Repair Complex Tfiih RAD4-33
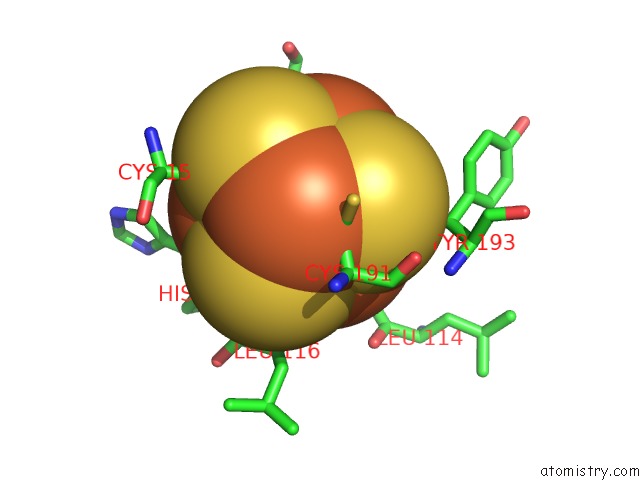
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Nucleotide Excision Repair Complex Tfiih RAD4-33 within 5.0Å range:
|
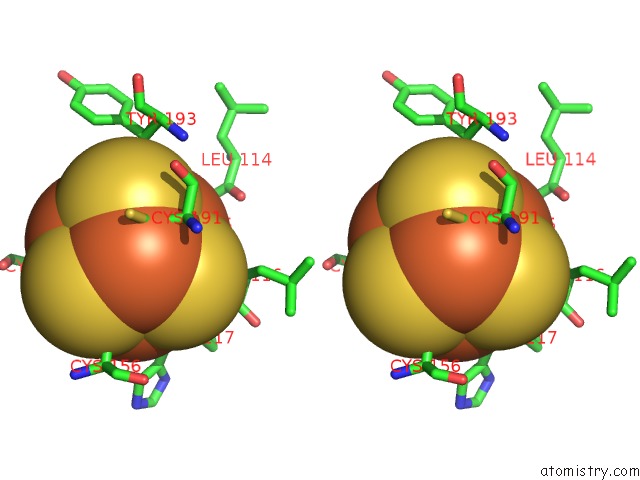
Iron binding site 2 out of 4 in 7m2u
Go back to
Iron binding site 2 out
of 4 in the Nucleotide Excision Repair Complex Tfiih RAD4-33
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Nucleotide Excision Repair Complex Tfiih RAD4-33 within 5.0Å range:
|
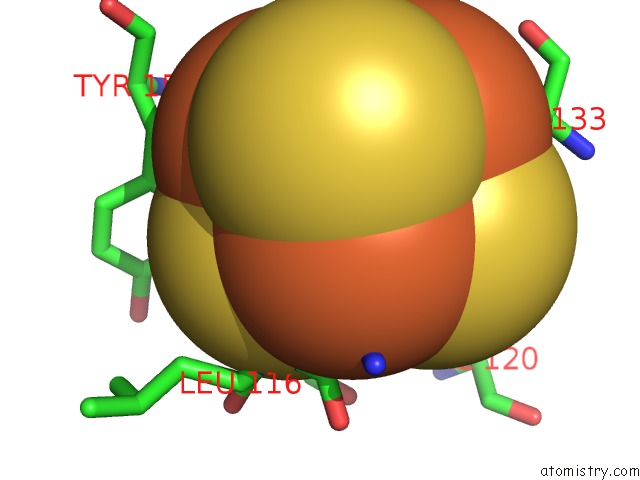
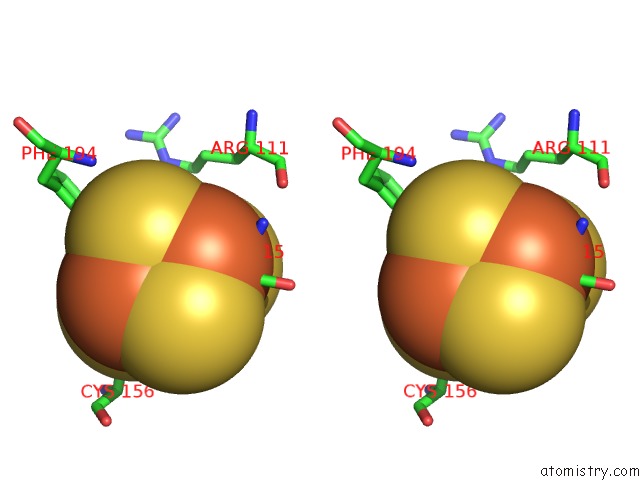
Iron binding site 3 out of 4 in 7m2u
Go back to
Iron binding site 3 out
of 4 in the Nucleotide Excision Repair Complex Tfiih RAD4-33
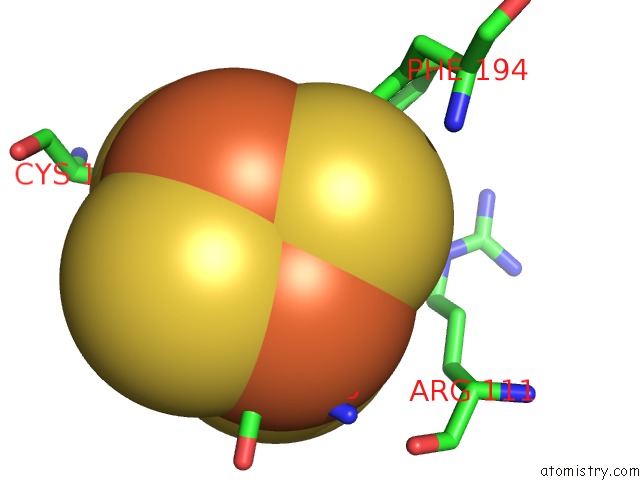
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 3 of Nucleotide Excision Repair Complex Tfiih RAD4-33 within 5.0Å range:
|
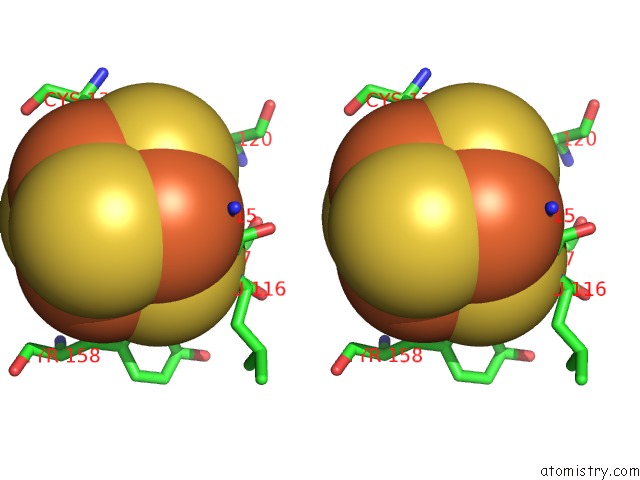
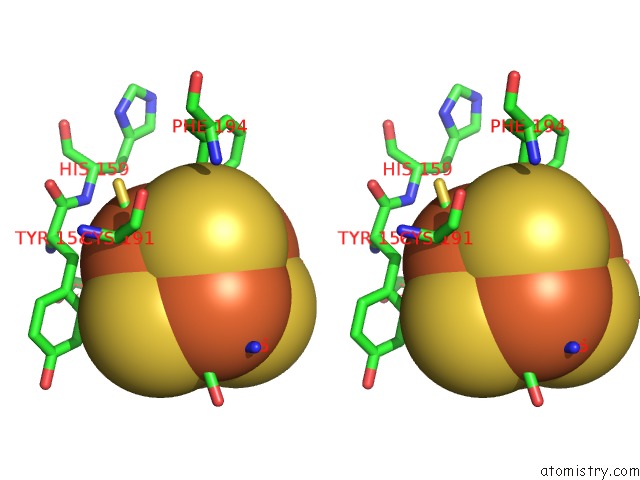
Iron binding site 4 out of 4 in 7m2u
Go back to
Iron binding site 4 out
of 4 in the Nucleotide Excision Repair Complex Tfiih RAD4-33
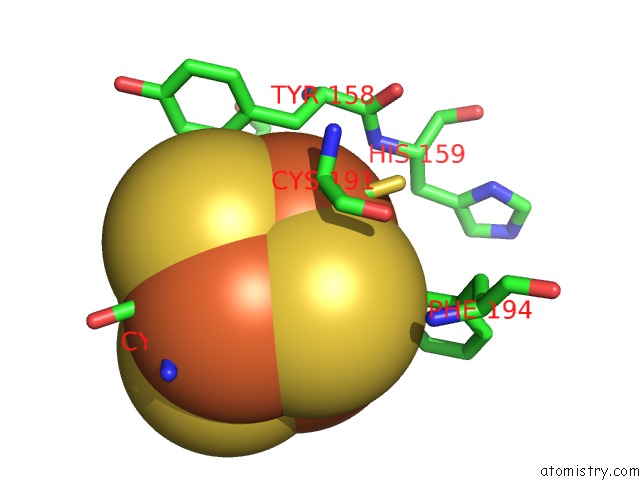
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 4 of Nucleotide Excision Repair Complex Tfiih RAD4-33 within 5.0Å range:
|
Reference:
T.Van Eeuwen,
Y.Shim,
H.J.Kim,
T.Zhao,
S.Basu,
B.A.Garcia,
C.D.Kaplan,
J.H.Min,
K.Murakami.
Cryo-Em Structure of Tfiih/RAD4-RAD23-RAD33 in Damaged Dna Opening in Nucleotide Excision Repair. Nat Commun V. 12 3338 2021.
ISSN: ESSN 2041-1723
PubMed: 34099686
DOI: 10.1038/S41467-021-23684-X
Page generated: Wed Aug 6 23:27:58 2025
ISSN: ESSN 2041-1723
PubMed: 34099686
DOI: 10.1038/S41467-021-23684-X
Last articles
K in 2FRZK in 2FPQ
K in 2FPM
K in 2FPL
K in 2FPK
K in 2FPG
K in 2FP4
K in 2FEU
K in 2FFY
K in 2FDE