Iron »
PDB 8css-8djt »
8dh6 »
Iron in PDB 8dh6: Cryo-Em Structure of Saccharomyces Cerevisiae Cytochrome C Oxidase (Complex IV) Extracted in Lipid Nanodiscs
Enzymatic activity of Cryo-Em Structure of Saccharomyces Cerevisiae Cytochrome C Oxidase (Complex IV) Extracted in Lipid Nanodiscs
All present enzymatic activity of Cryo-Em Structure of Saccharomyces Cerevisiae Cytochrome C Oxidase (Complex IV) Extracted in Lipid Nanodiscs:
7.1.1.9;
7.1.1.9;
Other elements in 8dh6:
The structure of Cryo-Em Structure of Saccharomyces Cerevisiae Cytochrome C Oxidase (Complex IV) Extracted in Lipid Nanodiscs also contains other interesting chemical elements:
| Zinc | (Zn) | 1 atom |
| Copper | (Cu) | 2 atoms |
| Calcium | (Ca) | 1 atom |
| Magnesium | (Mg) | 1 atom |
Iron Binding Sites:
The binding sites of Iron atom in the Cryo-Em Structure of Saccharomyces Cerevisiae Cytochrome C Oxidase (Complex IV) Extracted in Lipid Nanodiscs
(pdb code 8dh6). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 2 binding sites of Iron where determined in the Cryo-Em Structure of Saccharomyces Cerevisiae Cytochrome C Oxidase (Complex IV) Extracted in Lipid Nanodiscs, PDB code: 8dh6:
Jump to Iron binding site number: 1; 2;
In total 2 binding sites of Iron where determined in the Cryo-Em Structure of Saccharomyces Cerevisiae Cytochrome C Oxidase (Complex IV) Extracted in Lipid Nanodiscs, PDB code: 8dh6:
Jump to Iron binding site number: 1; 2;
Iron binding site 1 out of 2 in 8dh6
Go back to
Iron binding site 1 out
of 2 in the Cryo-Em Structure of Saccharomyces Cerevisiae Cytochrome C Oxidase (Complex IV) Extracted in Lipid Nanodiscs
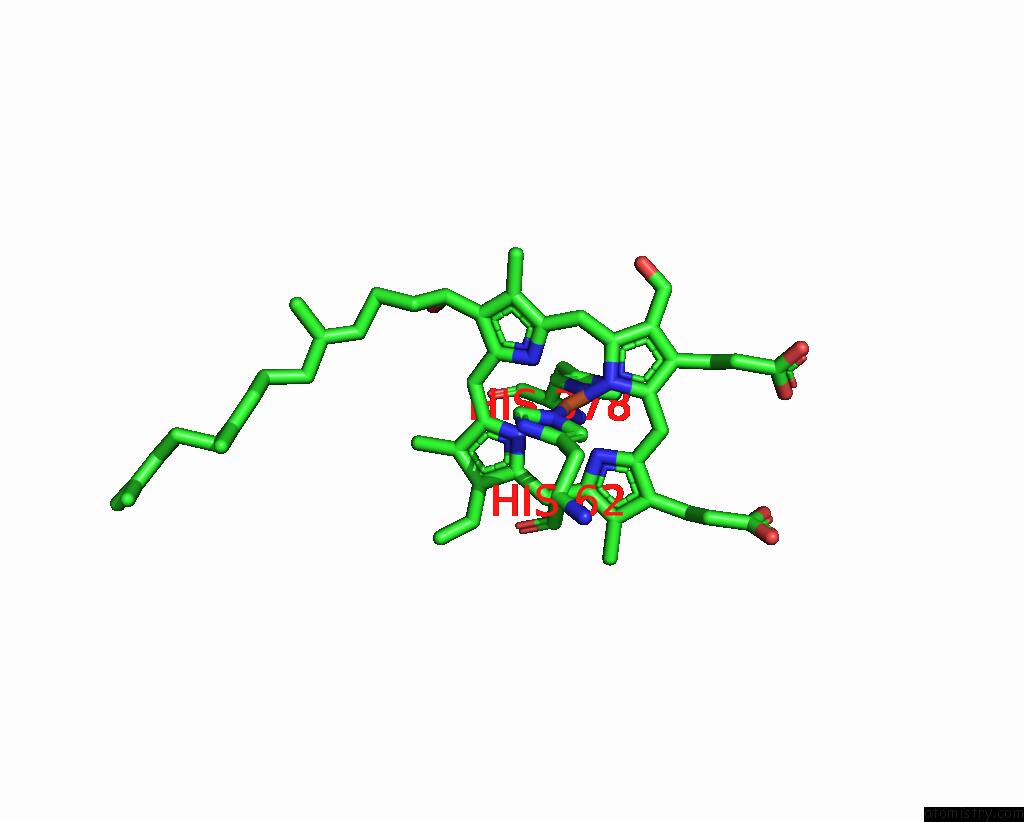
Mono view
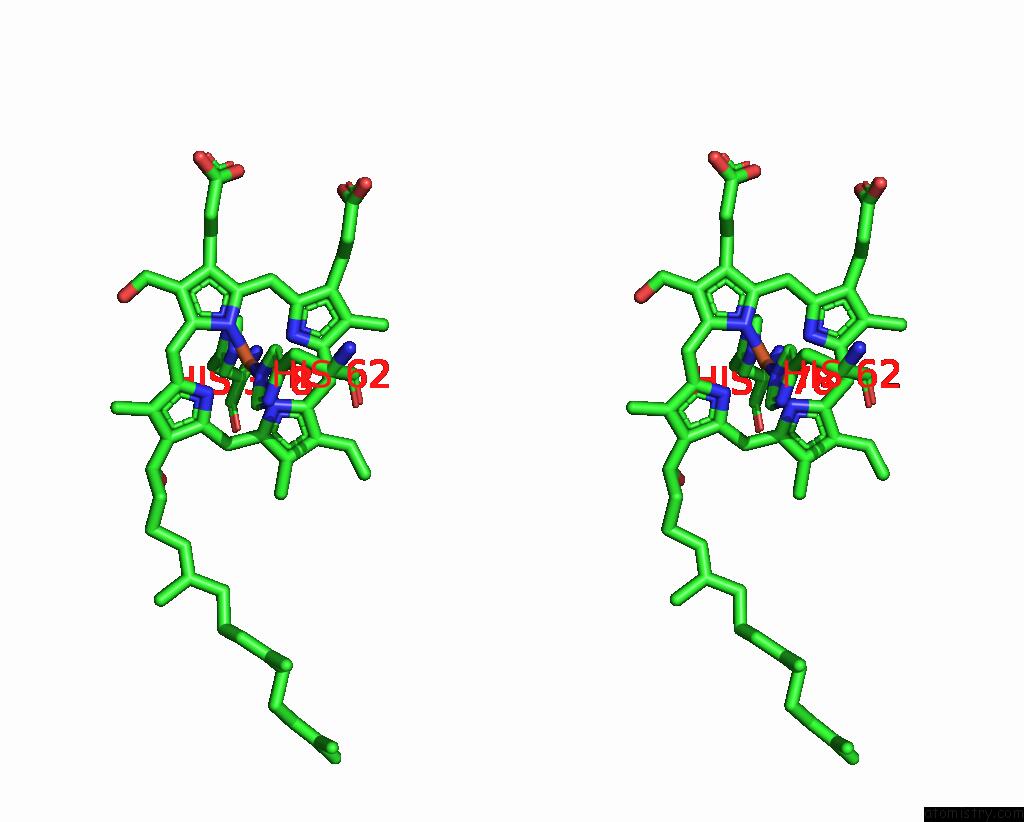
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Cryo-Em Structure of Saccharomyces Cerevisiae Cytochrome C Oxidase (Complex IV) Extracted in Lipid Nanodiscs within 5.0Å range:
|
Iron binding site 2 out of 2 in 8dh6
Go back to
Iron binding site 2 out
of 2 in the Cryo-Em Structure of Saccharomyces Cerevisiae Cytochrome C Oxidase (Complex IV) Extracted in Lipid Nanodiscs
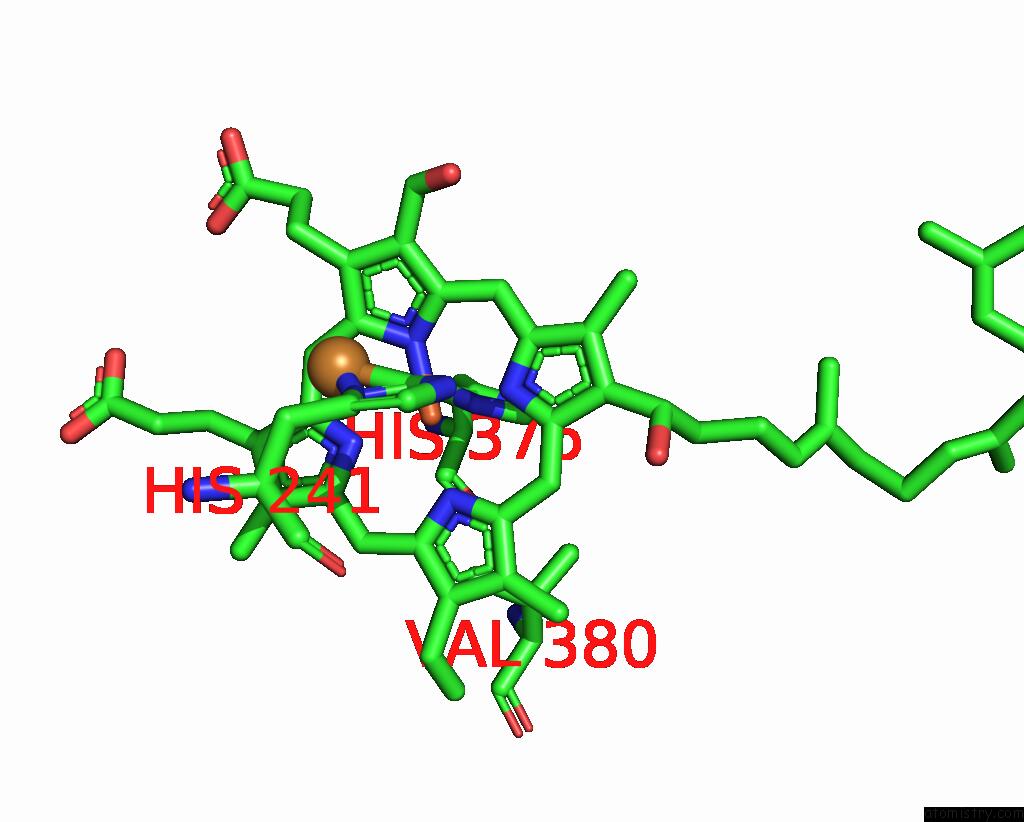
Mono view
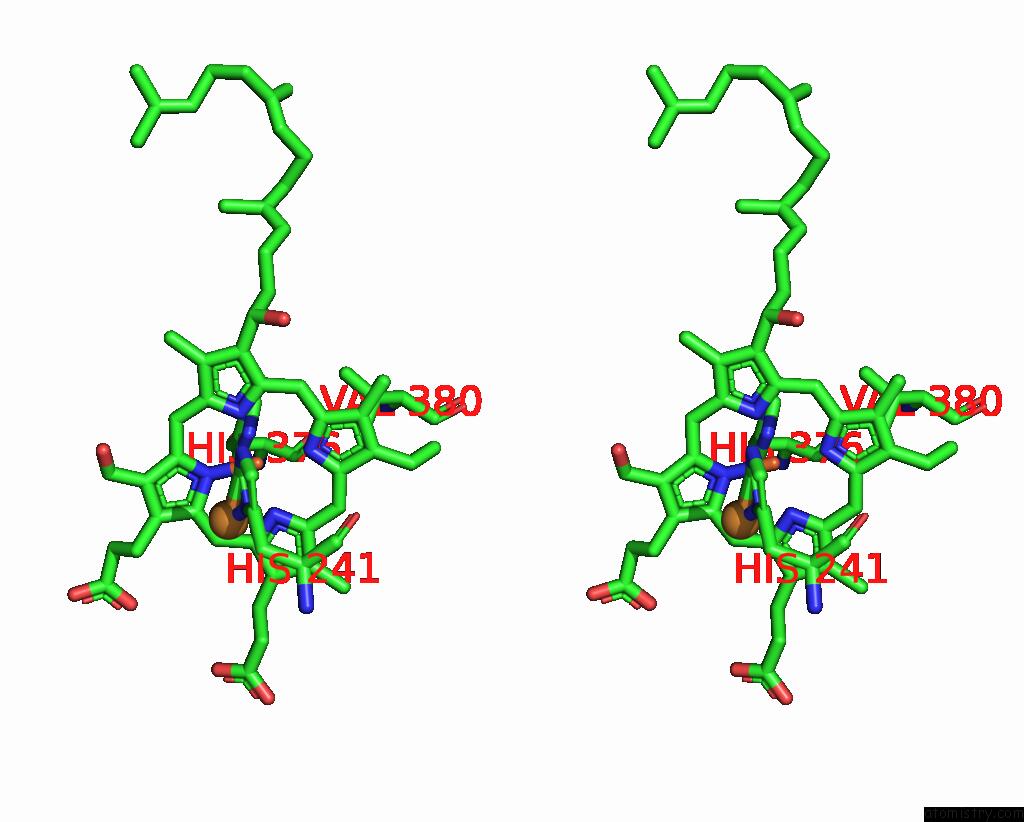
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Cryo-Em Structure of Saccharomyces Cerevisiae Cytochrome C Oxidase (Complex IV) Extracted in Lipid Nanodiscs within 5.0Å range:
|
Reference:
A.S.Godoy,
Y.Song,
H.Cheruvara,
A.Quigley,
G.Oliva.
Cryo-Em Structure of Saccharomyces Cerevisiae Cytochrome C Oxidase (Complex IV) Extracted in Lipid Nanodiscs To Be Published.
Page generated: Thu Aug 7 15:38:55 2025
Last articles
Mn in 2GU6Mn in 2GU5
Mn in 2GU4
Mn in 2FFL
Mn in 2GTX
Mn in 2GNM
Mn in 2GND
Mn in 2GMV
Mn in 2GLF
Mn in 2GLK