Iron »
PDB 8iba-8irf »
8iig »
Iron in PDB 8iig: Complex Form of Msmudgx H109A Mutant and Uracil- Obtained From Uracil Dna (Ttutt) Post Its Cleavage By Udgx H109A
Enzymatic activity of Complex Form of Msmudgx H109A Mutant and Uracil- Obtained From Uracil Dna (Ttutt) Post Its Cleavage By Udgx H109A
All present enzymatic activity of Complex Form of Msmudgx H109A Mutant and Uracil- Obtained From Uracil Dna (Ttutt) Post Its Cleavage By Udgx H109A:
3.2.2.27;
3.2.2.27;
Protein crystallography data
The structure of Complex Form of Msmudgx H109A Mutant and Uracil- Obtained From Uracil Dna (Ttutt) Post Its Cleavage By Udgx H109A, PDB code: 8iig
was solved by
S.Aroli,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 26.65 / 2.30 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 36.71, 50.72, 55.06, 90, 104.5, 90 |
| R / Rfree (%) | 18.6 / 22.2 |
Iron Binding Sites:
The binding sites of Iron atom in the Complex Form of Msmudgx H109A Mutant and Uracil- Obtained From Uracil Dna (Ttutt) Post Its Cleavage By Udgx H109A
(pdb code 8iig). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 4 binding sites of Iron where determined in the Complex Form of Msmudgx H109A Mutant and Uracil- Obtained From Uracil Dna (Ttutt) Post Its Cleavage By Udgx H109A, PDB code: 8iig:
Jump to Iron binding site number: 1; 2; 3; 4;
In total 4 binding sites of Iron where determined in the Complex Form of Msmudgx H109A Mutant and Uracil- Obtained From Uracil Dna (Ttutt) Post Its Cleavage By Udgx H109A, PDB code: 8iig:
Jump to Iron binding site number: 1; 2; 3; 4;
Iron binding site 1 out of 4 in 8iig
Go back to
Iron binding site 1 out
of 4 in the Complex Form of Msmudgx H109A Mutant and Uracil- Obtained From Uracil Dna (Ttutt) Post Its Cleavage By Udgx H109A
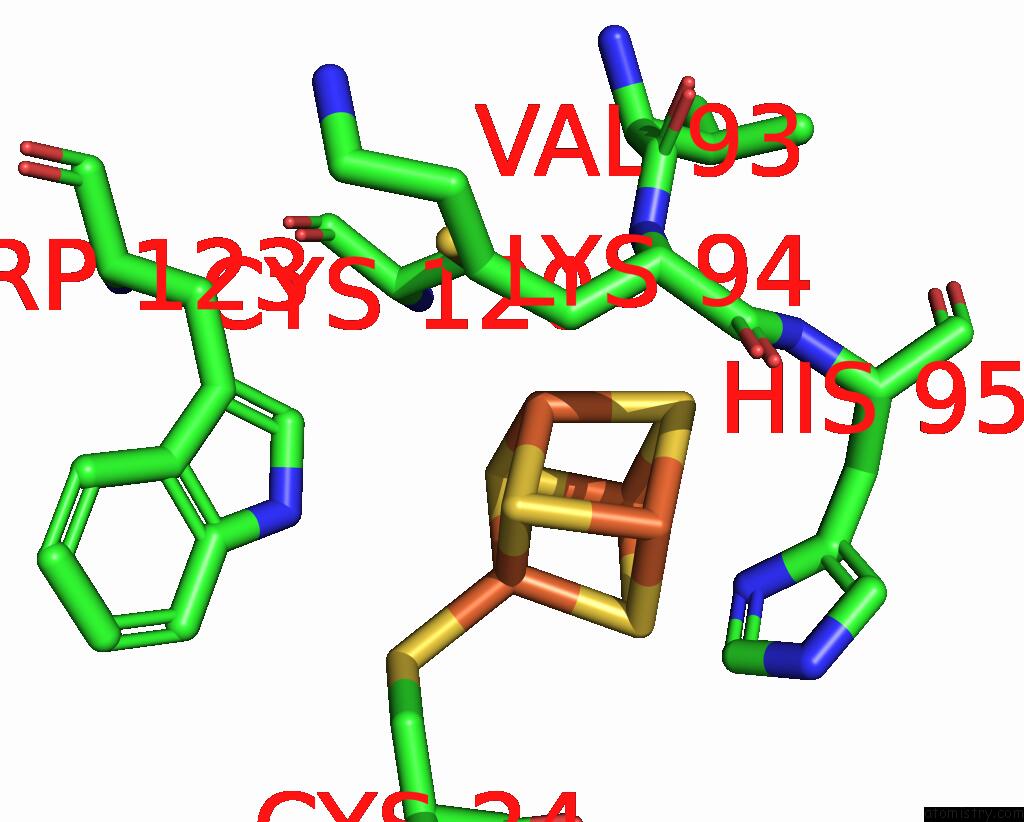
Mono view
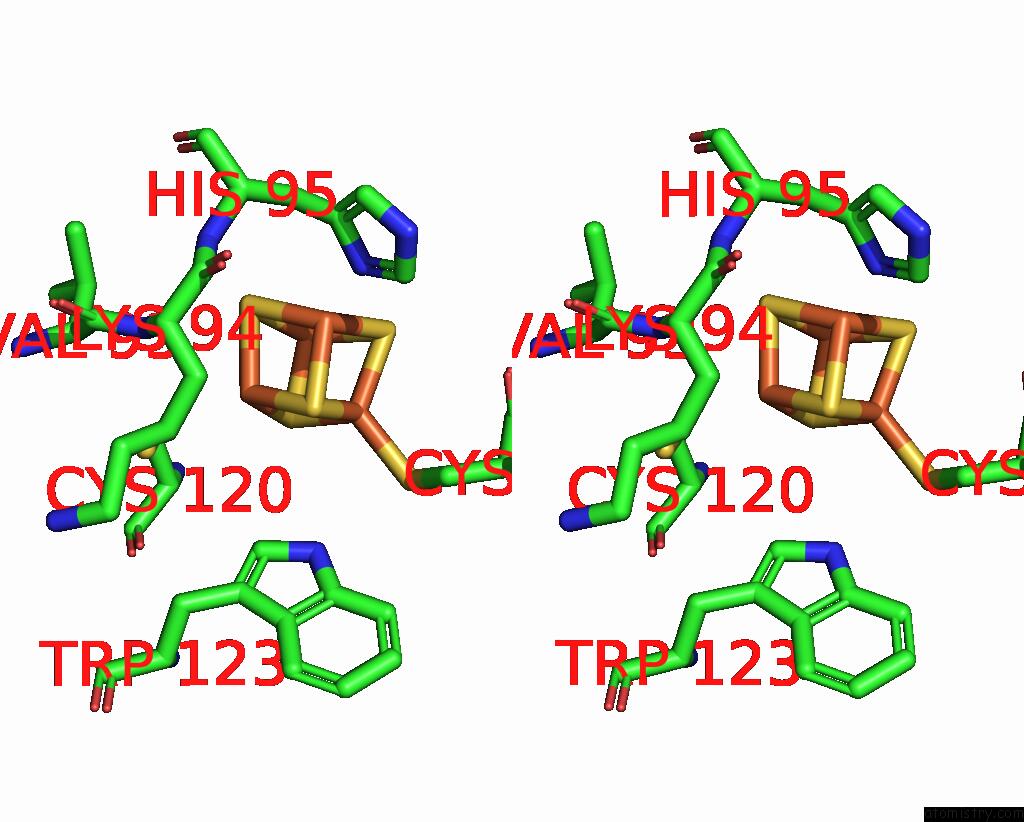
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Complex Form of Msmudgx H109A Mutant and Uracil- Obtained From Uracil Dna (Ttutt) Post Its Cleavage By Udgx H109A within 5.0Å range:
|
Iron binding site 2 out of 4 in 8iig
Go back to
Iron binding site 2 out
of 4 in the Complex Form of Msmudgx H109A Mutant and Uracil- Obtained From Uracil Dna (Ttutt) Post Its Cleavage By Udgx H109A
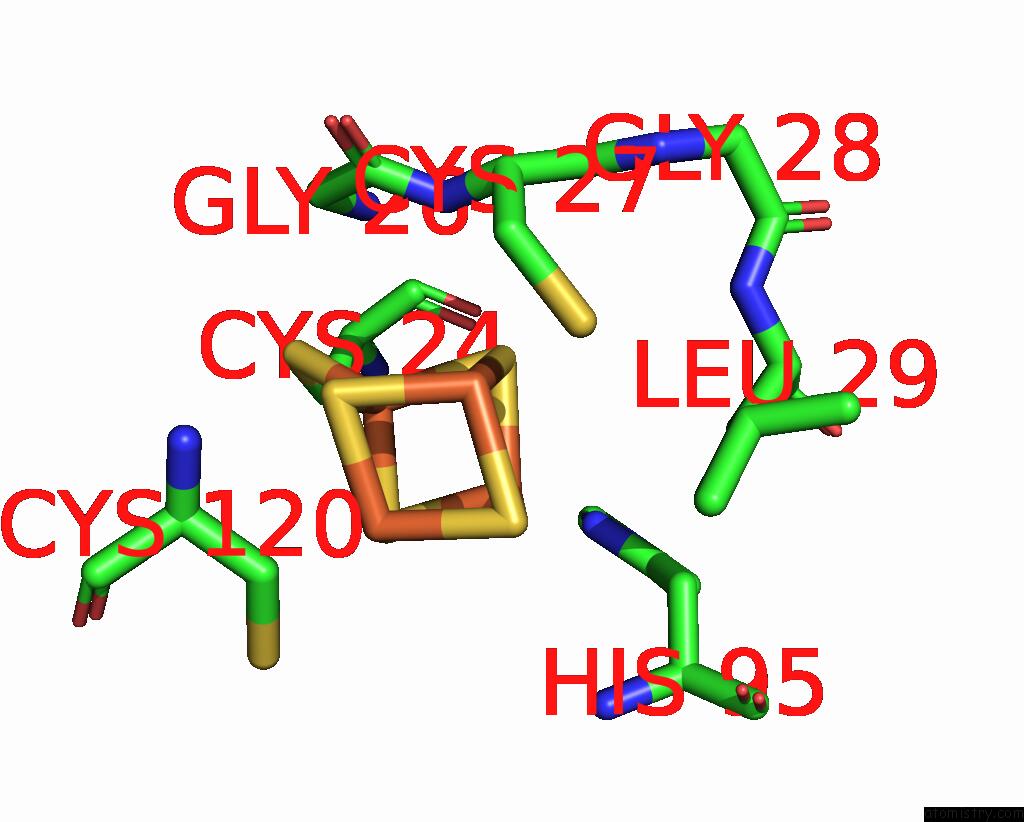
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Complex Form of Msmudgx H109A Mutant and Uracil- Obtained From Uracil Dna (Ttutt) Post Its Cleavage By Udgx H109A within 5.0Å range:
|
Iron binding site 3 out of 4 in 8iig
Go back to
Iron binding site 3 out
of 4 in the Complex Form of Msmudgx H109A Mutant and Uracil- Obtained From Uracil Dna (Ttutt) Post Its Cleavage By Udgx H109A
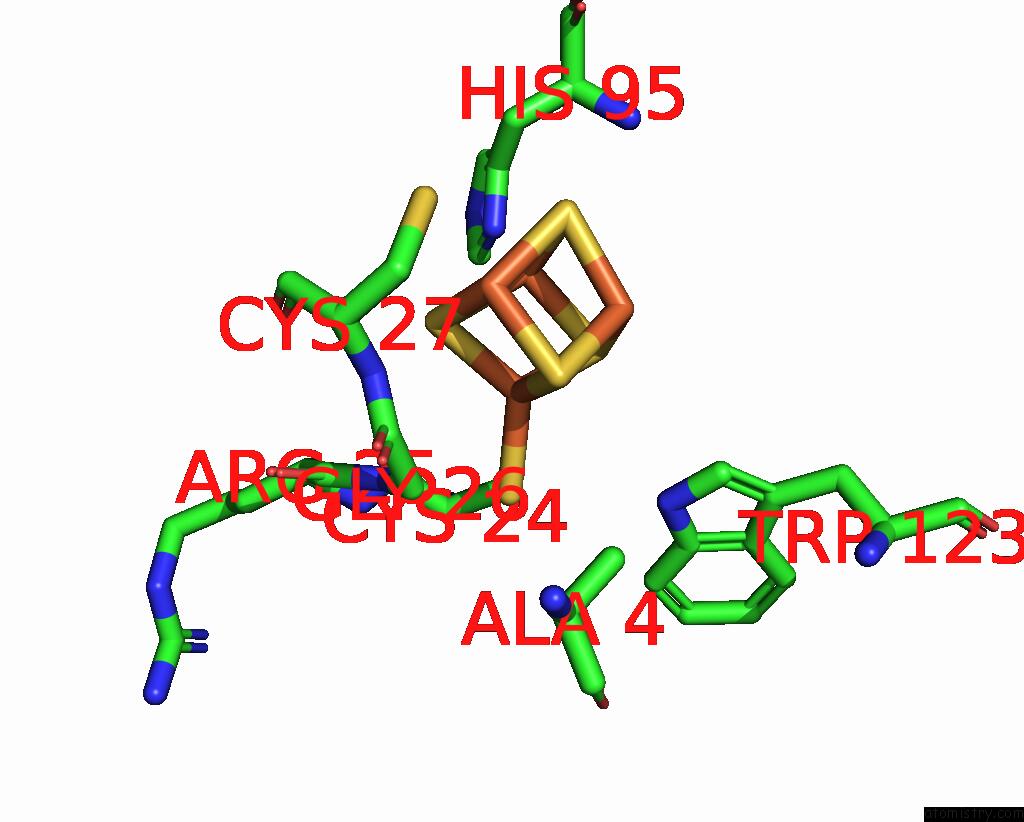
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 3 of Complex Form of Msmudgx H109A Mutant and Uracil- Obtained From Uracil Dna (Ttutt) Post Its Cleavage By Udgx H109A within 5.0Å range:
|
Iron binding site 4 out of 4 in 8iig
Go back to
Iron binding site 4 out
of 4 in the Complex Form of Msmudgx H109A Mutant and Uracil- Obtained From Uracil Dna (Ttutt) Post Its Cleavage By Udgx H109A

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 4 of Complex Form of Msmudgx H109A Mutant and Uracil- Obtained From Uracil Dna (Ttutt) Post Its Cleavage By Udgx H109A within 5.0Å range:
|
Reference:
S.Aroli,
E.J.Woo,
B.Gopal,
U.Varshney.
Mutational and Structural Analyses of Udgx: Insights Into the Active Site Pocket Architecture and Its Evolution. Nucleic Acids Res. 2023.
ISSN: ESSN 1362-4962
PubMed: 37283083
DOI: 10.1093/NAR/GKAD486
Page generated: Thu Aug 7 18:03:41 2025
ISSN: ESSN 1362-4962
PubMed: 37283083
DOI: 10.1093/NAR/GKAD486
Last articles
K in 9NESK in 9PHG
K in 9NEI
K in 9NED
K in 9NEC
K in 9NEG
K in 9CWU
K in 9CVB
K in 9CVA
K in 9COM