Iron »
PDB 9n5v-9vie »
9nqu »
Iron in PDB 9nqu: KDM6B-Nucleosome Structure Stabilized By H3K27C-UNC8015 Covalent Conjugate
Enzymatic activity of KDM6B-Nucleosome Structure Stabilized By H3K27C-UNC8015 Covalent Conjugate
All present enzymatic activity of KDM6B-Nucleosome Structure Stabilized By H3K27C-UNC8015 Covalent Conjugate:
1.14.11.68;
1.14.11.68;
Other elements in 9nqu:
The structure of KDM6B-Nucleosome Structure Stabilized By H3K27C-UNC8015 Covalent Conjugate also contains other interesting chemical elements:
| Zinc | (Zn) | 1 atom |
Iron Binding Sites:
The binding sites of Iron atom in the KDM6B-Nucleosome Structure Stabilized By H3K27C-UNC8015 Covalent Conjugate
(pdb code 9nqu). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total only one binding site of Iron was determined in the KDM6B-Nucleosome Structure Stabilized By H3K27C-UNC8015 Covalent Conjugate, PDB code: 9nqu:
In total only one binding site of Iron was determined in the KDM6B-Nucleosome Structure Stabilized By H3K27C-UNC8015 Covalent Conjugate, PDB code: 9nqu:
Iron binding site 1 out of 1 in 9nqu
Go back to
Iron binding site 1 out
of 1 in the KDM6B-Nucleosome Structure Stabilized By H3K27C-UNC8015 Covalent Conjugate
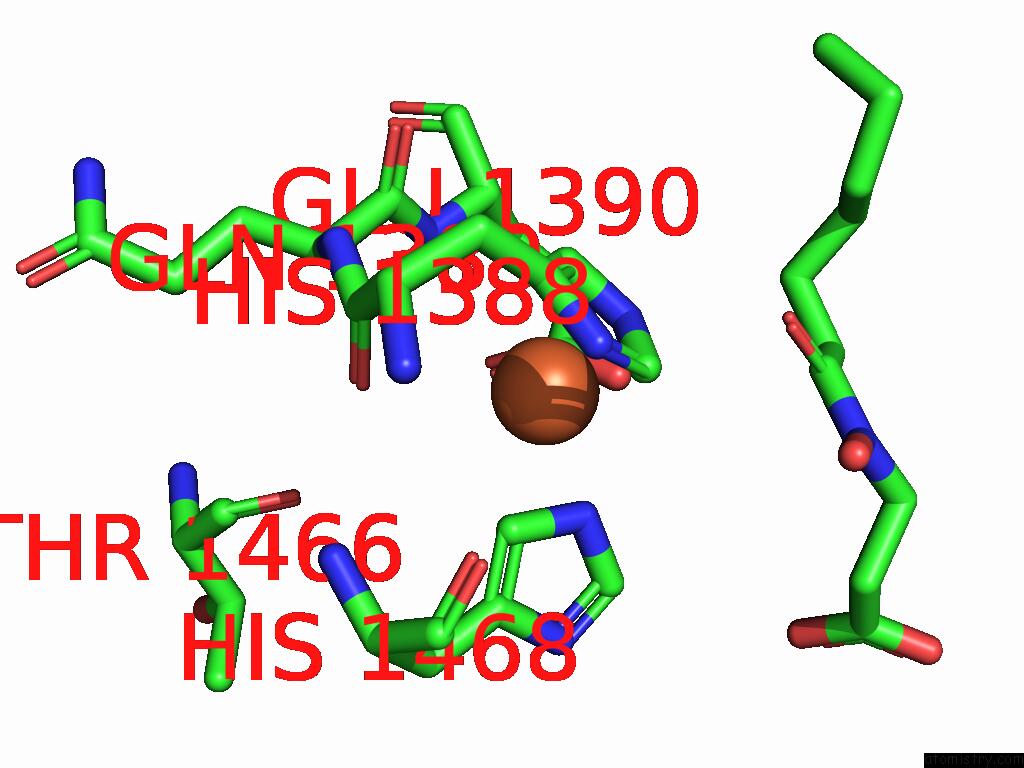
Mono view
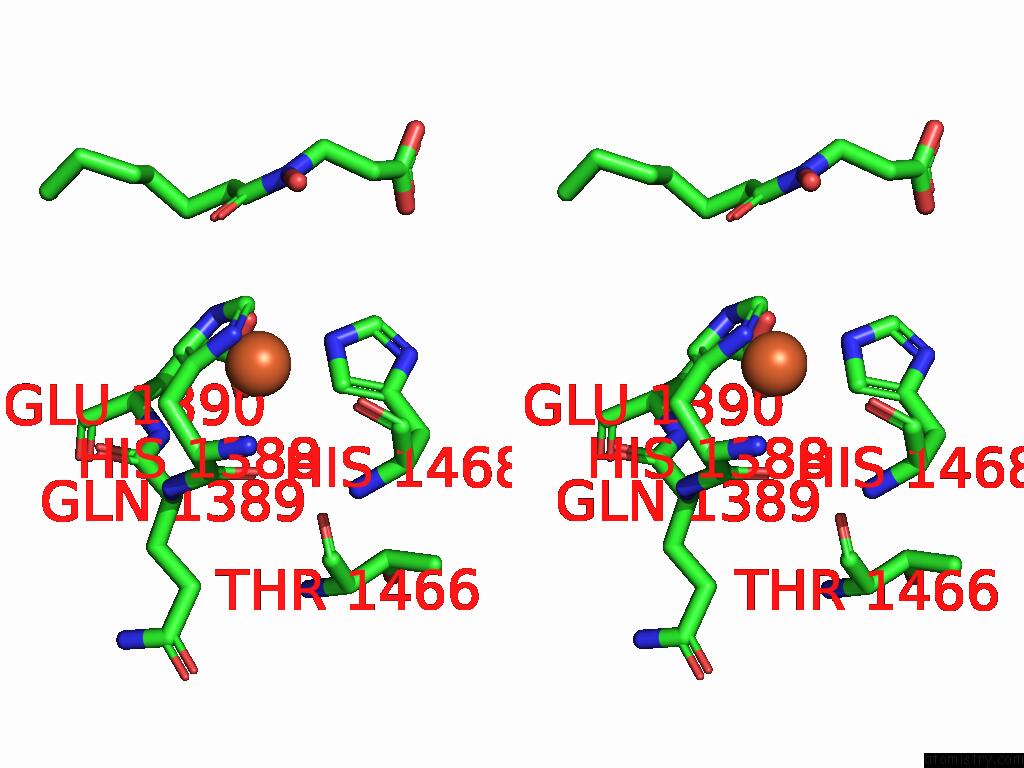
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of KDM6B-Nucleosome Structure Stabilized By H3K27C-UNC8015 Covalent Conjugate within 5.0Å range:
|
Reference:
C.-C.Lin,
R.K.Mcginty.
Structural Mechanism of H3K27 Demethylation and Crosstalk with Heterochromatin Markers Mol.Cell 2025.
ISSN: ISSN 1097-2765
Page generated: Fri Aug 8 07:55:46 2025
ISSN: ISSN 1097-2765
Last articles
Hg in 6RH4Hg in 6RG5
Hg in 6RFH
Hg in 6PII
Hg in 6LNH
Hg in 6NJX
Hg in 6KR6
Hg in 6IQV
Hg in 6L8O
Hg in 6BZI