Iron »
PDB 1fz2-1gek »
1gej »
Iron in PDB 1gej: Structural Characterization of N-Butyl-Isocyanide Complexes of Cytochromes P450NOR and P450CAM
Protein crystallography data
The structure of Structural Characterization of N-Butyl-Isocyanide Complexes of Cytochromes P450NOR and P450CAM, PDB code: 1gej
was solved by
D.-S.Lee,
S.-Y.Park,
K.Yamane,
Y.Shiro,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 25.00 / 1.50 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 54.590, 81.910, 85.830, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.7 / 24.1 |
Iron Binding Sites:
The binding sites of Iron atom in the Structural Characterization of N-Butyl-Isocyanide Complexes of Cytochromes P450NOR and P450CAM
(pdb code 1gej). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total only one binding site of Iron was determined in the Structural Characterization of N-Butyl-Isocyanide Complexes of Cytochromes P450NOR and P450CAM, PDB code: 1gej:
In total only one binding site of Iron was determined in the Structural Characterization of N-Butyl-Isocyanide Complexes of Cytochromes P450NOR and P450CAM, PDB code: 1gej:
Iron binding site 1 out of 1 in 1gej
Go back to
Iron binding site 1 out
of 1 in the Structural Characterization of N-Butyl-Isocyanide Complexes of Cytochromes P450NOR and P450CAM
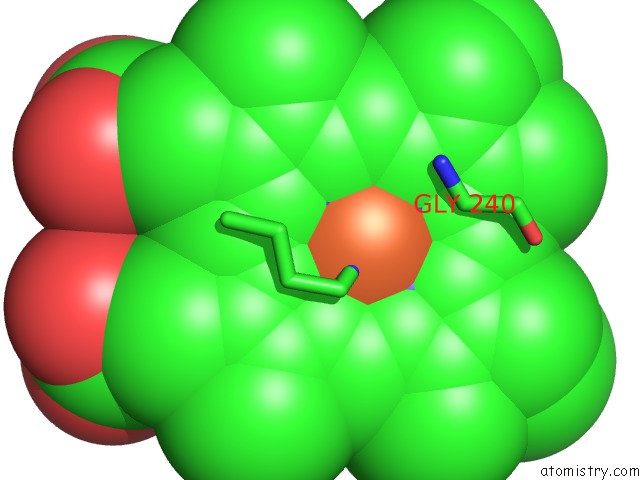
Mono view
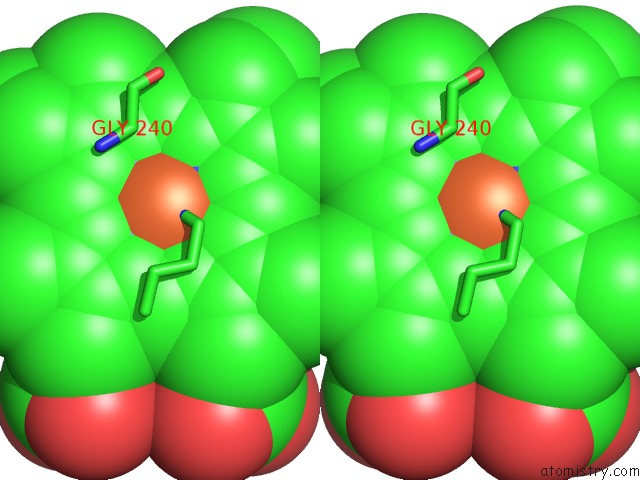
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Structural Characterization of N-Butyl-Isocyanide Complexes of Cytochromes P450NOR and P450CAM within 5.0Å range:
|
Reference:
D.S.Lee,
S.Y.Park,
K.Yamane,
E.Obayashi,
H.Hori,
Y.Shiro.
Structural Characterization of N-Butyl-Isocyanide Complexes of Cytochromes P450NOR and P450CAM. Biochemistry V. 40 2669 2001.
ISSN: ISSN 0006-2960
PubMed: 11258878
DOI: 10.1021/BI002225S
Page generated: Sat Aug 3 05:57:06 2024
ISSN: ISSN 0006-2960
PubMed: 11258878
DOI: 10.1021/BI002225S
Last articles
Cl in 5WL7Cl in 5WLG
Cl in 5WL5
Cl in 5WKX
Cl in 5WKY
Cl in 5WL1
Cl in 5WHU
Cl in 5WKV
Cl in 5WKR
Cl in 5WKU