Iron »
PDB 1iro-1j3y »
1izo »
Iron in PDB 1izo: Cytochrome P450 Bs Beta Complexed with Fatty Acid
Protein crystallography data
The structure of Cytochrome P450 Bs Beta Complexed with Fatty Acid, PDB code: 1izo
was solved by
D.S.Lee,
A.Yamada,
H.Sugimoto,
I.Matsunaga,
H.Ogura,
K.Ichihara,
S.Adachi,
S.Y.Park,
Y.Shiro,
Riken Structuralgenomics/Proteomics Initiative (Rsgi),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.00 / 2.10 |
| Space group | P 32 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 155.789, 155.789, 111.904, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 24.6 / 28 |
Iron Binding Sites:
The binding sites of Iron atom in the Cytochrome P450 Bs Beta Complexed with Fatty Acid
(pdb code 1izo). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 3 binding sites of Iron where determined in the Cytochrome P450 Bs Beta Complexed with Fatty Acid, PDB code: 1izo:
Jump to Iron binding site number: 1; 2; 3;
In total 3 binding sites of Iron where determined in the Cytochrome P450 Bs Beta Complexed with Fatty Acid, PDB code: 1izo:
Jump to Iron binding site number: 1; 2; 3;
Iron binding site 1 out of 3 in 1izo
Go back to
Iron binding site 1 out
of 3 in the Cytochrome P450 Bs Beta Complexed with Fatty Acid
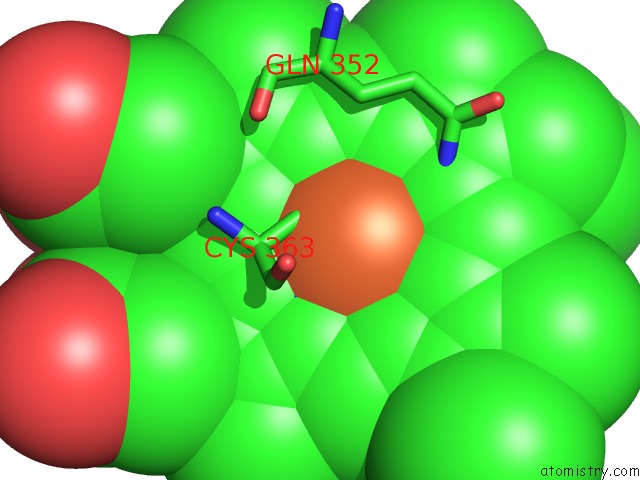
Mono view
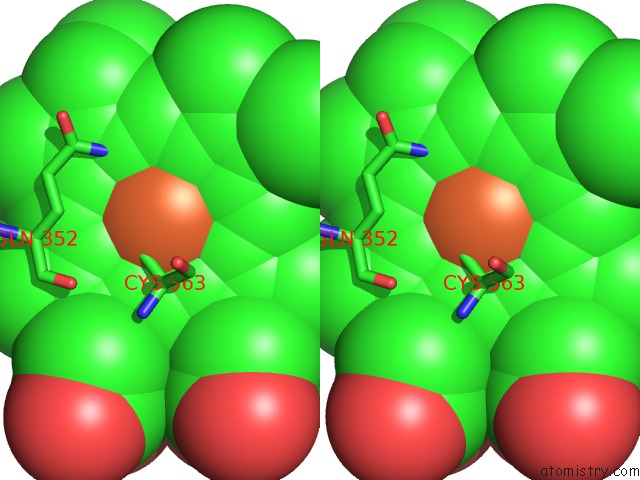
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Cytochrome P450 Bs Beta Complexed with Fatty Acid within 5.0Å range:
|
Iron binding site 2 out of 3 in 1izo
Go back to
Iron binding site 2 out
of 3 in the Cytochrome P450 Bs Beta Complexed with Fatty Acid
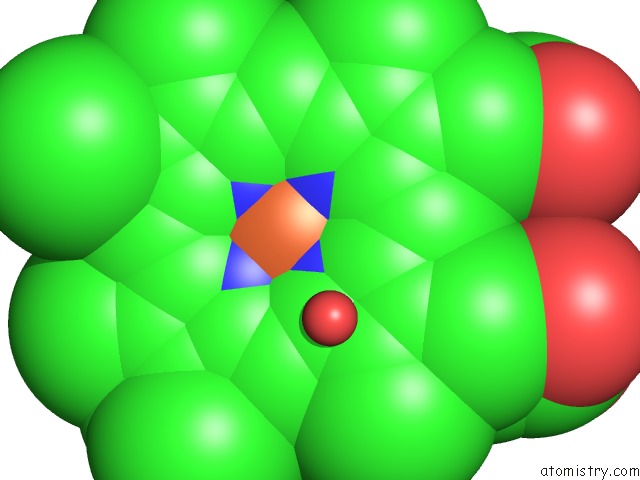
Mono view
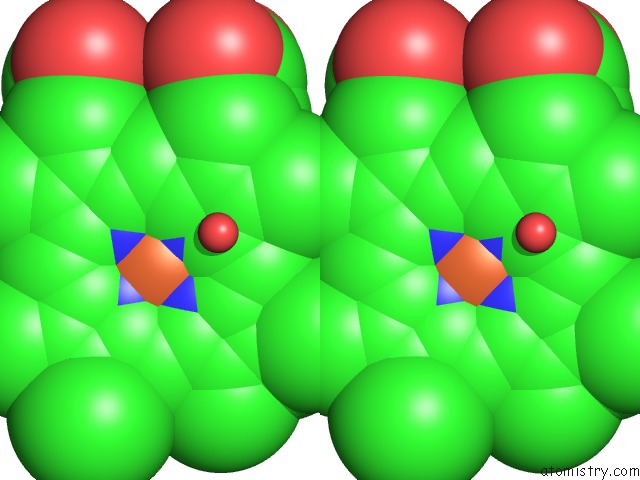
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Cytochrome P450 Bs Beta Complexed with Fatty Acid within 5.0Å range:
|
Iron binding site 3 out of 3 in 1izo
Go back to
Iron binding site 3 out
of 3 in the Cytochrome P450 Bs Beta Complexed with Fatty Acid
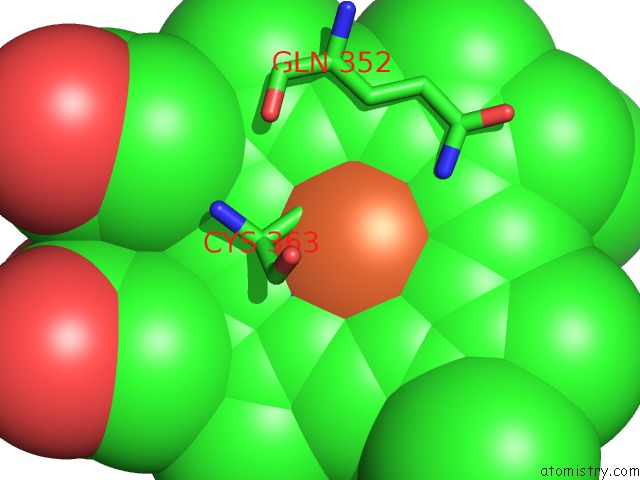
Mono view
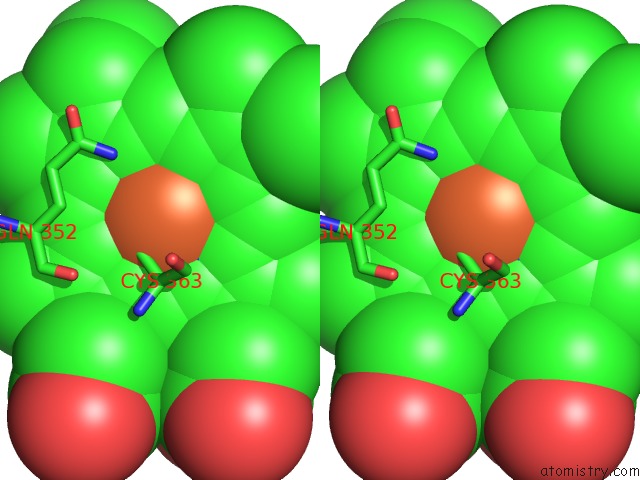
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 3 of Cytochrome P450 Bs Beta Complexed with Fatty Acid within 5.0Å range:
|
Reference:
D.S.Lee,
A.Yamada,
H.Sugimoto,
I.Matsunaga,
H.Ogura,
K.Ichihara,
S.Adachi,
S.Y.Park,
Y.Shiro.
Substrate Recognition and Molecular Mechanism of Fatty Acid Hydroxylation By Cytochrome P450 From Bacillus Subtilis. Crystallographic, Spectroscopic, and Mutational Studies. J.Biol.Chem. V. 278 9761 2003.
ISSN: ISSN 0021-9258
PubMed: 12519760
DOI: 10.1074/JBC.M211575200
Page generated: Sat Aug 3 08:18:32 2024
ISSN: ISSN 0021-9258
PubMed: 12519760
DOI: 10.1074/JBC.M211575200
Last articles
Cl in 5WGTCl in 5WGR
Cl in 5WG9
Cl in 5WFZ
Cl in 5WFQ
Cl in 5WFW
Cl in 5WFP
Cl in 5WFM
Cl in 5WEO
Cl in 5WFO