Iron »
PDB 1k2r-1kqg »
1k3g »
Iron in PDB 1k3g: uc(Nmr) Solution Structure of Oxidized Cytochrome C-553 From Bacillus Pasteurii
Iron Binding Sites:
The binding sites of Iron atom in the uc(Nmr) Solution Structure of Oxidized Cytochrome C-553 From Bacillus Pasteurii
(pdb code 1k3g). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total only one binding site of Iron was determined in the uc(Nmr) Solution Structure of Oxidized Cytochrome C-553 From Bacillus Pasteurii, PDB code: 1k3g:
In total only one binding site of Iron was determined in the uc(Nmr) Solution Structure of Oxidized Cytochrome C-553 From Bacillus Pasteurii, PDB code: 1k3g:
Iron binding site 1 out of 1 in 1k3g
Go back to
Iron binding site 1 out
of 1 in the uc(Nmr) Solution Structure of Oxidized Cytochrome C-553 From Bacillus Pasteurii
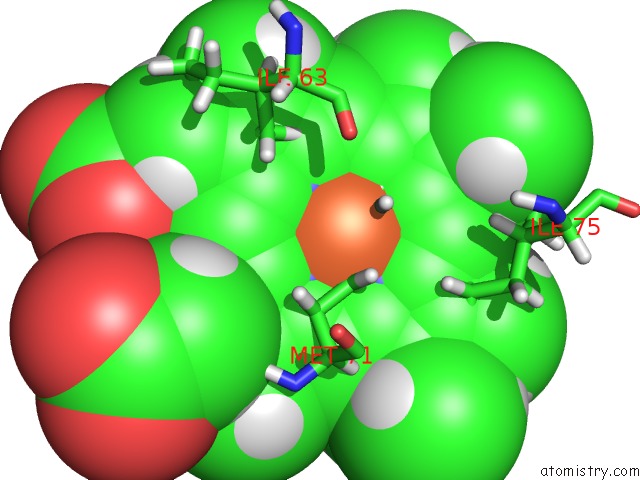
Mono view
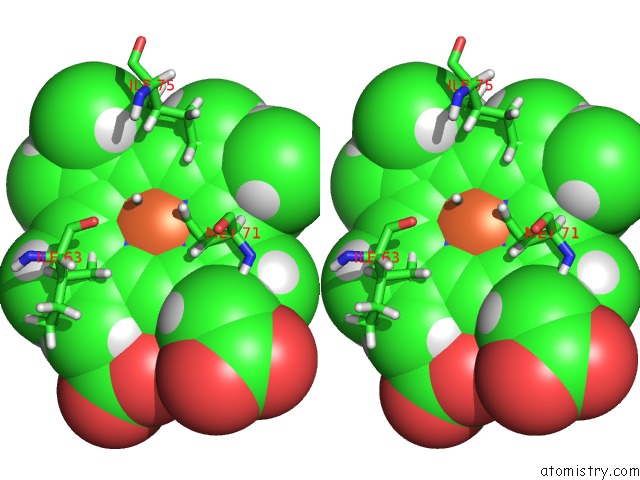
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of uc(Nmr) Solution Structure of Oxidized Cytochrome C-553 From Bacillus Pasteurii within 5.0Å range:
|
Reference:
L.Banci,
I.Bertini,
S.Ciurli,
A.Dikiy,
J.Dittmer,
A.Rosato,
G.Sciara,
A.R.Thompsett.
uc(Nmr) Solution Structure, Backbone Mobility, and Homology Modeling of C-Type Cytochromes From Gram-Positive Bacteria. Chembiochem V. 3 299 2002.
ISSN: ISSN 1439-4227
PubMed: 11933230
DOI: 10.1002/1439-7633(20020402)3:4<299::AID-CBIC299>3.0.CO;2-0
Page generated: Wed Jul 16 16:56:02 2025
ISSN: ISSN 1439-4227
PubMed: 11933230
DOI: 10.1002/1439-7633(20020402)3:4<299::AID-CBIC299>3.0.CO;2-0
Last articles
Fe in 2YXOFe in 2YRS
Fe in 2YXC
Fe in 2YNM
Fe in 2YVJ
Fe in 2YP1
Fe in 2YU2
Fe in 2YU1
Fe in 2YQB
Fe in 2YOO