Iron »
PDB 1mpy-1n5u »
1mqv »
Iron in PDB 1mqv: Crystal Structure of the Q1A/F32W/W72F Mutant of Rhodopseudomonas Palustris Cytochrome C' (Prime) Expressed in E. Coli
Protein crystallography data
The structure of Crystal Structure of the Q1A/F32W/W72F Mutant of Rhodopseudomonas Palustris Cytochrome C' (Prime) Expressed in E. Coli, PDB code: 1mqv
was solved by
J.C.Lee,
K.C.Engman,
F.A.Tezcan,
H.B.Gray,
J.R.Winkler,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 42.00 / 1.78 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 33.839, 62.085, 113.419, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 21.4 / 26.5 |
Iron Binding Sites:
The binding sites of Iron atom in the Crystal Structure of the Q1A/F32W/W72F Mutant of Rhodopseudomonas Palustris Cytochrome C' (Prime) Expressed in E. Coli
(pdb code 1mqv). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 2 binding sites of Iron where determined in the Crystal Structure of the Q1A/F32W/W72F Mutant of Rhodopseudomonas Palustris Cytochrome C' (Prime) Expressed in E. Coli, PDB code: 1mqv:
Jump to Iron binding site number: 1; 2;
In total 2 binding sites of Iron where determined in the Crystal Structure of the Q1A/F32W/W72F Mutant of Rhodopseudomonas Palustris Cytochrome C' (Prime) Expressed in E. Coli, PDB code: 1mqv:
Jump to Iron binding site number: 1; 2;
Iron binding site 1 out of 2 in 1mqv
Go back to
Iron binding site 1 out
of 2 in the Crystal Structure of the Q1A/F32W/W72F Mutant of Rhodopseudomonas Palustris Cytochrome C' (Prime) Expressed in E. Coli
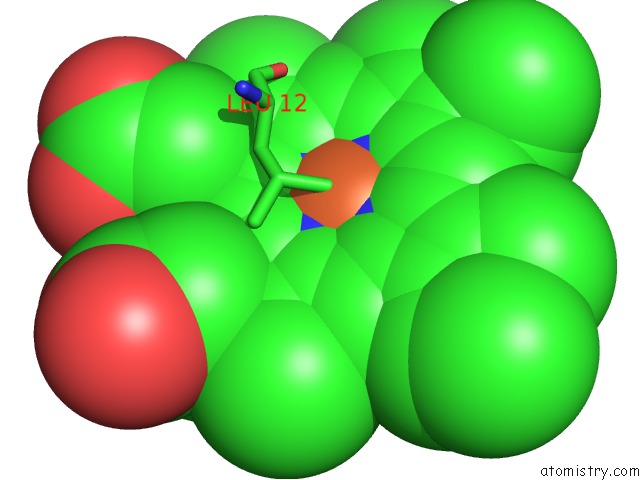
Mono view
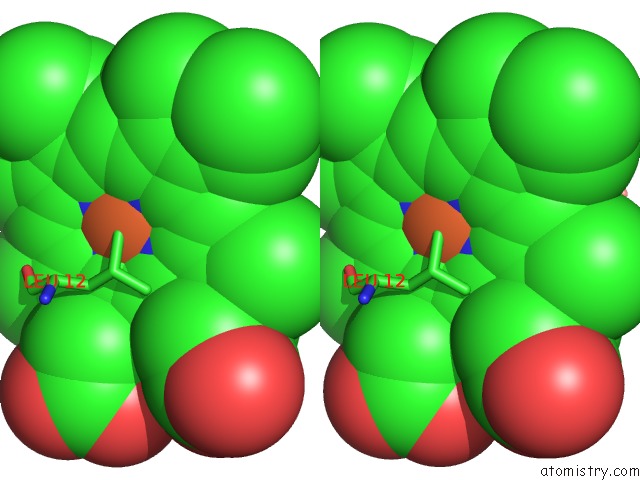
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Crystal Structure of the Q1A/F32W/W72F Mutant of Rhodopseudomonas Palustris Cytochrome C' (Prime) Expressed in E. Coli within 5.0Å range:
|
Iron binding site 2 out of 2 in 1mqv
Go back to
Iron binding site 2 out
of 2 in the Crystal Structure of the Q1A/F32W/W72F Mutant of Rhodopseudomonas Palustris Cytochrome C' (Prime) Expressed in E. Coli
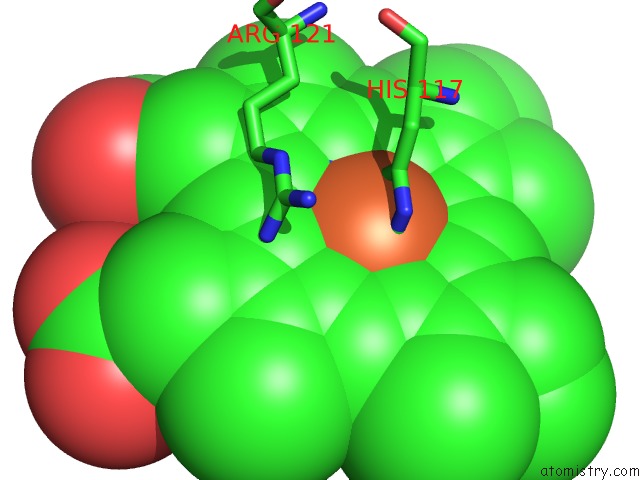
Mono view
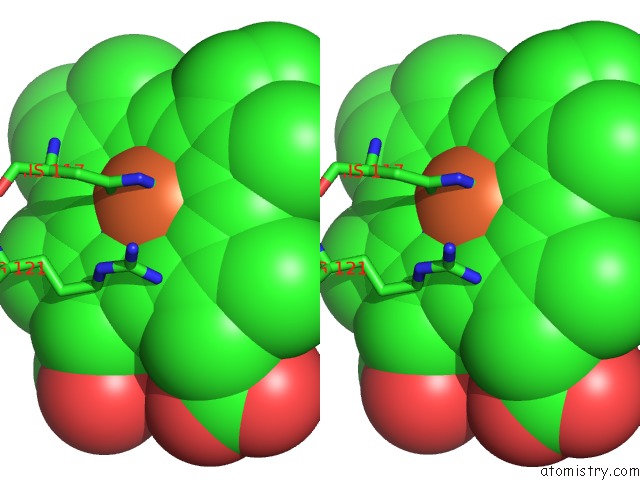
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Crystal Structure of the Q1A/F32W/W72F Mutant of Rhodopseudomonas Palustris Cytochrome C' (Prime) Expressed in E. Coli within 5.0Å range:
|
Reference:
J.C.Lee,
K.C.Engman,
F.A.Tezcan,
H.B.Gray,
J.R.Winkler.
Structural Features of Cytochrome C' Folding Intermediates Revealed By Fluorescence Energy-Transfer Kinetics Proc.Natl.Acad.Sci.Usa V. 99 14778 2002.
ISSN: ISSN 0027-8424
PubMed: 12407175
DOI: 10.1073/PNAS.192574099
Page generated: Sat Aug 3 11:07:34 2024
ISSN: ISSN 0027-8424
PubMed: 12407175
DOI: 10.1073/PNAS.192574099
Last articles
F in 4Q09F in 4PKO
F in 4PKN
F in 4Q06
F in 4Q02
F in 4PYY
F in 4PZH
F in 4PYX
F in 4PX5
F in 4PY4