Iron »
PDB 1ozr-1pha »
1p6n »
Iron in PDB 1p6n: Bovine Endothelial Nos Heme Domain with L-N(Omega)-Nitroarginine-(4R)- Amino-L-Proline Amide Bound
Enzymatic activity of Bovine Endothelial Nos Heme Domain with L-N(Omega)-Nitroarginine-(4R)- Amino-L-Proline Amide Bound
All present enzymatic activity of Bovine Endothelial Nos Heme Domain with L-N(Omega)-Nitroarginine-(4R)- Amino-L-Proline Amide Bound:
1.14.13.39;
1.14.13.39;
Protein crystallography data
The structure of Bovine Endothelial Nos Heme Domain with L-N(Omega)-Nitroarginine-(4R)- Amino-L-Proline Amide Bound, PDB code: 1p6n
was solved by
M.L.Flinspach,
H.Li,
J.Jamal,
W.Yang,
H.Huang,
J.-M.Hah,
J.A.Gomez-Vidal,
E.A.Litzinger,
R.B.Silverman,
T.L.Poulos,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 42.55 / 2.50 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 57.669, 106.292, 156.524, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 21.8 / 27.8 |
Other elements in 1p6n:
The structure of Bovine Endothelial Nos Heme Domain with L-N(Omega)-Nitroarginine-(4R)- Amino-L-Proline Amide Bound also contains other interesting chemical elements:
| Arsenic | (As) | 2 atoms |
| Zinc | (Zn) | 1 atom |
Iron Binding Sites:
The binding sites of Iron atom in the Bovine Endothelial Nos Heme Domain with L-N(Omega)-Nitroarginine-(4R)- Amino-L-Proline Amide Bound
(pdb code 1p6n). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 2 binding sites of Iron where determined in the Bovine Endothelial Nos Heme Domain with L-N(Omega)-Nitroarginine-(4R)- Amino-L-Proline Amide Bound, PDB code: 1p6n:
Jump to Iron binding site number: 1; 2;
In total 2 binding sites of Iron where determined in the Bovine Endothelial Nos Heme Domain with L-N(Omega)-Nitroarginine-(4R)- Amino-L-Proline Amide Bound, PDB code: 1p6n:
Jump to Iron binding site number: 1; 2;
Iron binding site 1 out of 2 in 1p6n
Go back to
Iron binding site 1 out
of 2 in the Bovine Endothelial Nos Heme Domain with L-N(Omega)-Nitroarginine-(4R)- Amino-L-Proline Amide Bound

Mono view
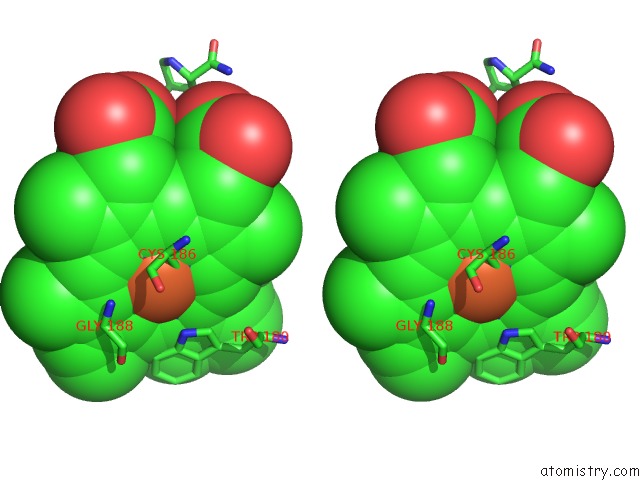
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Bovine Endothelial Nos Heme Domain with L-N(Omega)-Nitroarginine-(4R)- Amino-L-Proline Amide Bound within 5.0Å range:
|
Iron binding site 2 out of 2 in 1p6n
Go back to
Iron binding site 2 out
of 2 in the Bovine Endothelial Nos Heme Domain with L-N(Omega)-Nitroarginine-(4R)- Amino-L-Proline Amide Bound

Mono view
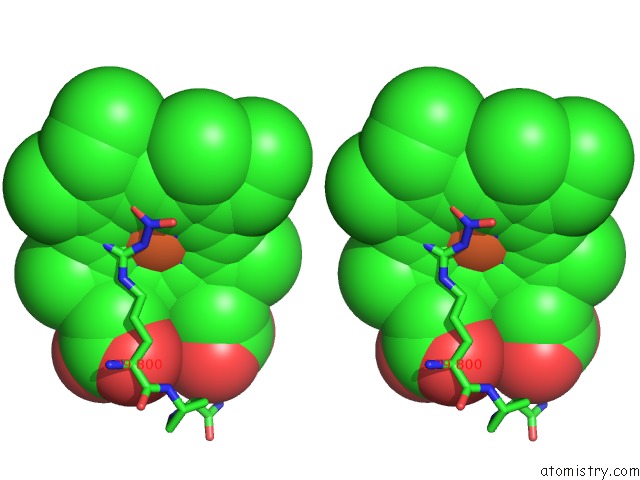
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Bovine Endothelial Nos Heme Domain with L-N(Omega)-Nitroarginine-(4R)- Amino-L-Proline Amide Bound within 5.0Å range:
|
Reference:
M.L.Flinspach,
H.Li,
J.Jamal,
W.Yang,
H.Huang,
J.M.Hah,
J.A.Gomez-Vidal,
E.A.Litzinger,
R.B.Silverman,
T.L.Poulos.
Structural Basis For Dipeptide Amide Isoform-Selective Inhibition of Neuronal Nitric Oxide Synthase. Nat.Struct.Mol.Biol. V. 11 54 2004.
ISSN: ISSN 1545-9993
PubMed: 14718923
DOI: 10.1038/NSMB704
Page generated: Sat Aug 3 12:59:06 2024
ISSN: ISSN 1545-9993
PubMed: 14718923
DOI: 10.1038/NSMB704
Last articles
F in 4TW9F in 4TYI
F in 4TY9
F in 4TXN
F in 4TVJ
F in 4TS2
F in 4TS0
F in 4TKG
F in 4TN4
F in 4TN6