Iron »
PDB 2d3y-2e1q »
2dkk »
Iron in PDB 2dkk: Structure/Function Studies of Cytochrome P450 158A1 From Streptomyces Coelicolor A3(2)
Protein crystallography data
The structure of Structure/Function Studies of Cytochrome P450 158A1 From Streptomyces Coelicolor A3(2), PDB code: 2dkk
was solved by
B.Zhao,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 7.99 / 1.97 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 103.968, 44.352, 102.107, 90.00, 114.44, 90.00 |
| R / Rfree (%) | 22.8 / 28.5 |
Iron Binding Sites:
The binding sites of Iron atom in the Structure/Function Studies of Cytochrome P450 158A1 From Streptomyces Coelicolor A3(2)
(pdb code 2dkk). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total only one binding site of Iron was determined in the Structure/Function Studies of Cytochrome P450 158A1 From Streptomyces Coelicolor A3(2), PDB code: 2dkk:
In total only one binding site of Iron was determined in the Structure/Function Studies of Cytochrome P450 158A1 From Streptomyces Coelicolor A3(2), PDB code: 2dkk:
Iron binding site 1 out of 1 in 2dkk
Go back to
Iron binding site 1 out
of 1 in the Structure/Function Studies of Cytochrome P450 158A1 From Streptomyces Coelicolor A3(2)
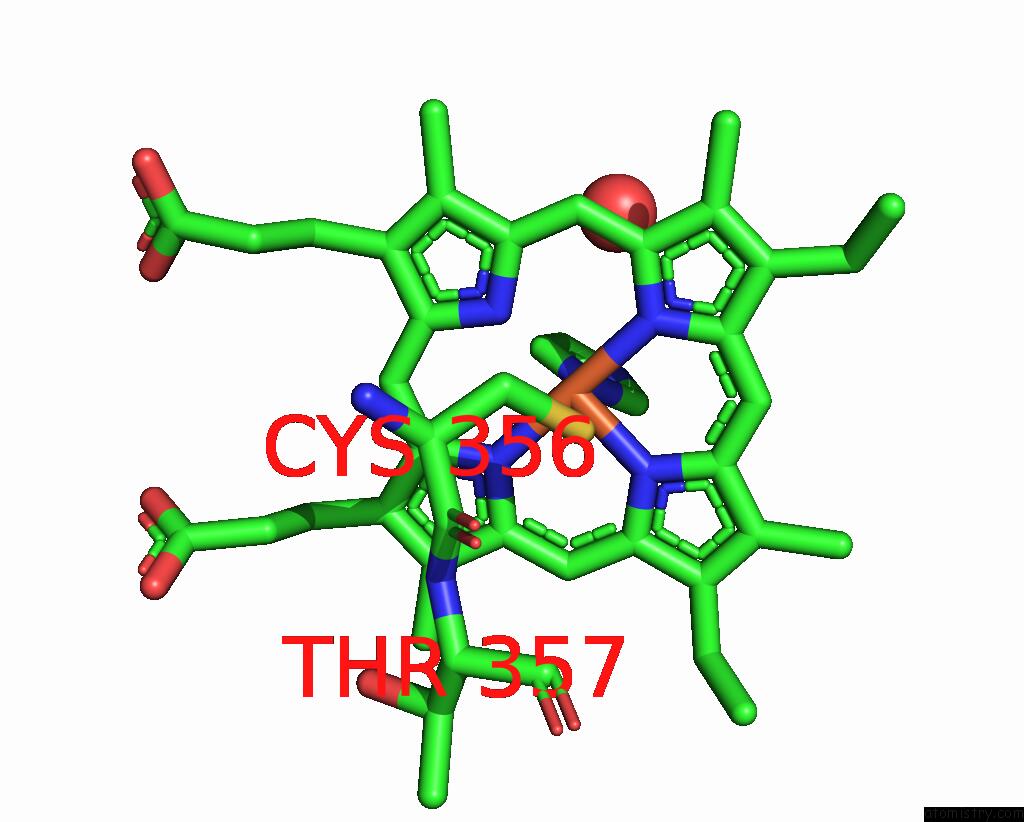
Mono view
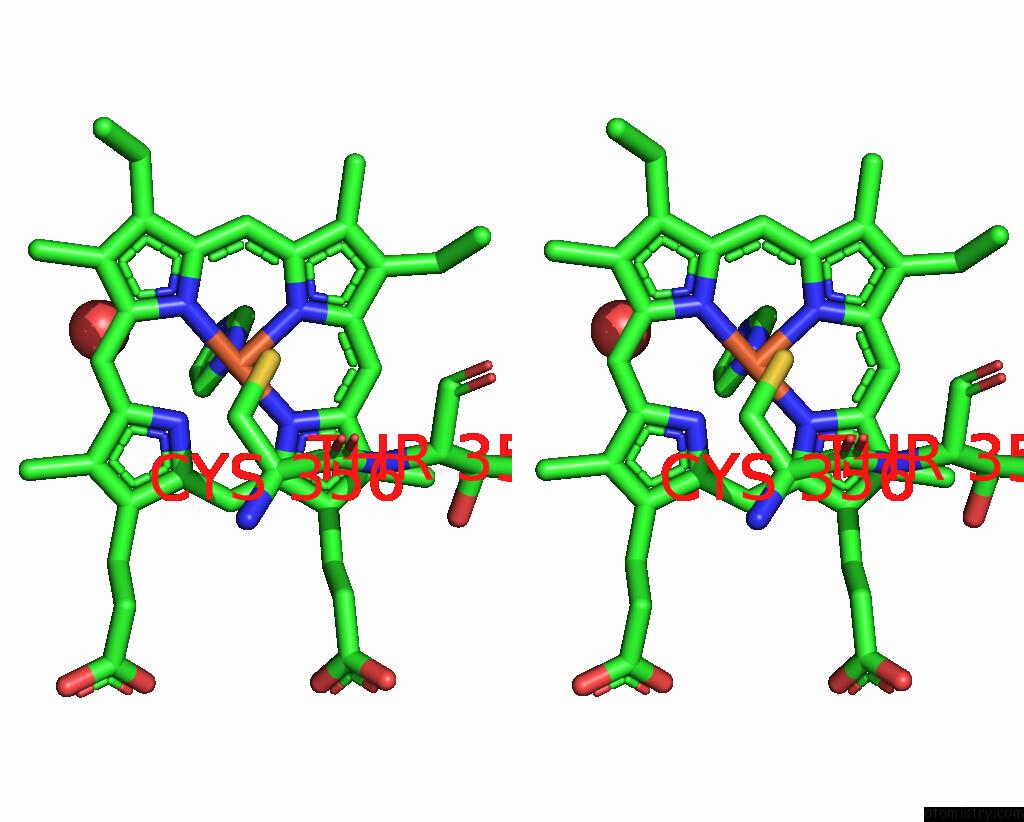
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Structure/Function Studies of Cytochrome P450 158A1 From Streptomyces Coelicolor A3(2) within 5.0Å range:
|
Reference:
B.Zhao,
D.C.Lamb,
L.Lei,
S.L.Kelly,
H.Yuan,
D.L.Hachey,
M.R.Waterman.
Different Binding Modes of Two Flaviolin Substrate Molecules in Cytochrome P450 158A1 (CYP158A1) Compared to CYP158A2. Biochemistry V. 46 8725 2007.
ISSN: ISSN 0006-2960
PubMed: 17614370
DOI: 10.1021/BI7006959
Page generated: Sat Aug 3 20:47:31 2024
ISSN: ISSN 0006-2960
PubMed: 17614370
DOI: 10.1021/BI7006959
Last articles
Cl in 5T03Cl in 5T02
Cl in 5T05
Cl in 5T09
Cl in 5SZD
Cl in 5SZV
Cl in 5SZZ
Cl in 5SZY
Cl in 5SYW
Cl in 5SYX