Iron »
PDB 2eut-2fkz »
2evp »
Iron in PDB 2evp: The Structures of Thiolate- and Carboxylate-Ligated Ferric H93G Myoglobin: Models For Cytochrome P450 and For Oxyanion-Bound Heme Proteins
Protein crystallography data
The structure of The Structures of Thiolate- and Carboxylate-Ligated Ferric H93G Myoglobin: Models For Cytochrome P450 and For Oxyanion-Bound Heme Proteins, PDB code: 2evp
was solved by
J.Qin,
R.Perera,
L.L.Lovelace,
J.H.Dawson,
L.Lebioda,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 1.70 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 39.670, 47.770, 77.430, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19.4 / 21.4 |
Iron Binding Sites:
The binding sites of Iron atom in the The Structures of Thiolate- and Carboxylate-Ligated Ferric H93G Myoglobin: Models For Cytochrome P450 and For Oxyanion-Bound Heme Proteins
(pdb code 2evp). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total only one binding site of Iron was determined in the The Structures of Thiolate- and Carboxylate-Ligated Ferric H93G Myoglobin: Models For Cytochrome P450 and For Oxyanion-Bound Heme Proteins, PDB code: 2evp:
In total only one binding site of Iron was determined in the The Structures of Thiolate- and Carboxylate-Ligated Ferric H93G Myoglobin: Models For Cytochrome P450 and For Oxyanion-Bound Heme Proteins, PDB code: 2evp:
Iron binding site 1 out of 1 in 2evp
Go back to
Iron binding site 1 out
of 1 in the The Structures of Thiolate- and Carboxylate-Ligated Ferric H93G Myoglobin: Models For Cytochrome P450 and For Oxyanion-Bound Heme Proteins
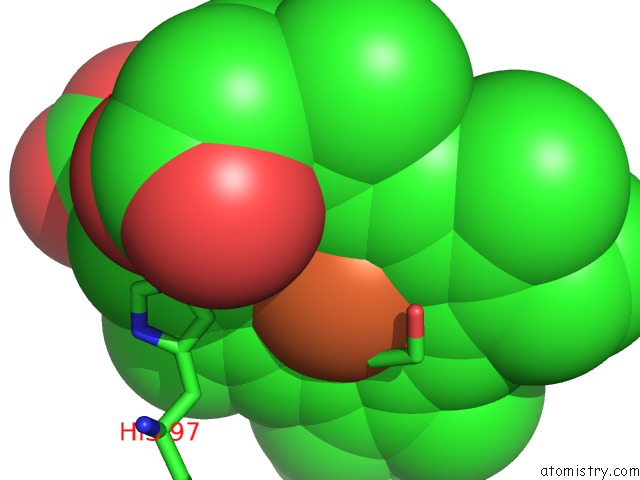
Mono view
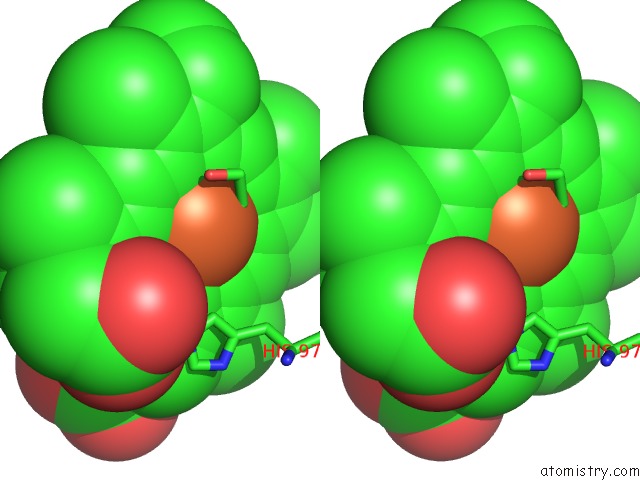
Stereo pair view

Mono view

Stereo pair view
|
|
A full contact list of Iron with other atoms in the Fe binding
site number 1 of The Structures of Thiolate- and Carboxylate-Ligated Ferric H93G Myoglobin: Models For Cytochrome P450 and For Oxyanion-Bound Heme Proteins within 5.0Å range:
|
Reference:
J.Qin,
R.Perera,
L.L.Lovelace,
J.H.Dawson,
L.Lebioda.
Structures of Thiolate- and Carboxylate-Ligated Ferric H93G Myoglobin: Models For Cytochrome P450 and For Oxyanion-Bound Heme Proteins(,). Biochemistry V. 45 3170 2006.
ISSN: ISSN 0006-2960
PubMed: 16519512
DOI: 10.1021/BI052171S
Page generated: Thu Jul 17 00:56:31 2025
ISSN: ISSN 0006-2960
PubMed: 16519512
DOI: 10.1021/BI052171S
Last articles
Zn in 8PSSZn in 8PSQ
Zn in 8PSO
Zn in 8PRC
Zn in 8PRF
Zn in 8PRE
Zn in 8PRB
Zn in 8PP7
Zn in 8PR4
Zn in 8PPV