Iron »
PDB 2mya-2nwb »
2n3y »
Iron in PDB 2n3y: uc(Nmr) Structure of the Y48PCMF Variant of Human Cytochrome C in Its Reduced State
Iron Binding Sites:
The binding sites of Iron atom in the uc(Nmr) Structure of the Y48PCMF Variant of Human Cytochrome C in Its Reduced State
(pdb code 2n3y). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total only one binding site of Iron was determined in the uc(Nmr) Structure of the Y48PCMF Variant of Human Cytochrome C in Its Reduced State, PDB code: 2n3y:
In total only one binding site of Iron was determined in the uc(Nmr) Structure of the Y48PCMF Variant of Human Cytochrome C in Its Reduced State, PDB code: 2n3y:
Iron binding site 1 out of 1 in 2n3y
Go back to
Iron binding site 1 out
of 1 in the uc(Nmr) Structure of the Y48PCMF Variant of Human Cytochrome C in Its Reduced State
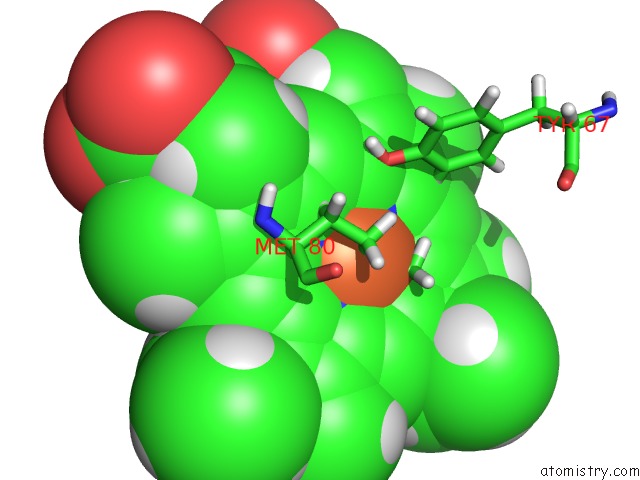
Mono view
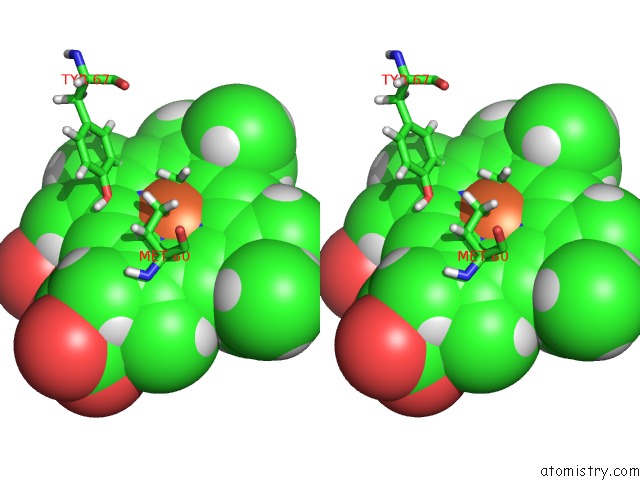
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of uc(Nmr) Structure of the Y48PCMF Variant of Human Cytochrome C in Its Reduced State within 5.0Å range:
|
Reference:
B.Moreno-Beltran,
A.Guerra-Castellano,
A.Diaz-Quintana,
R.Del Conte,
S.M.Garcia-Maurino,
S.Diaz-Moreno,
K.Gonzalez-Arzola,
C.Santos-Ocana,
A.Velazquez-Campoy,
M.A.De La Rosa,
P.Turano,
I.Diaz-Moreno.
Structural Basis of Mitochondrial Dysfunction in Response to Cytochrome C Phosphorylation at Tyrosine 48. Proc. Natl. Acad. Sci. V. 114 E3041 2017U.S.A..
ISSN: ESSN 1091-6490
PubMed: 28348229
DOI: 10.1073/PNAS.1618008114
Page generated: Thu Jul 17 03:00:02 2025
ISSN: ESSN 1091-6490
PubMed: 28348229
DOI: 10.1073/PNAS.1618008114
Last articles
Fe in 2YXOFe in 2YRS
Fe in 2YXC
Fe in 2YNM
Fe in 2YVJ
Fe in 2YP1
Fe in 2YU2
Fe in 2YU1
Fe in 2YQB
Fe in 2YOO