Iron »
PDB 3c25-3crb »
3cli »
Iron in PDB 3cli: Crystal Structure of Arabidopsis Thaliana Allene Oxide Synthase (Aos, Cytochrome P450 74A, CYP74A) at 1.80 A Resolution
Enzymatic activity of Crystal Structure of Arabidopsis Thaliana Allene Oxide Synthase (Aos, Cytochrome P450 74A, CYP74A) at 1.80 A Resolution
All present enzymatic activity of Crystal Structure of Arabidopsis Thaliana Allene Oxide Synthase (Aos, Cytochrome P450 74A, CYP74A) at 1.80 A Resolution:
4.2.1.92;
4.2.1.92;
Protein crystallography data
The structure of Crystal Structure of Arabidopsis Thaliana Allene Oxide Synthase (Aos, Cytochrome P450 74A, CYP74A) at 1.80 A Resolution, PDB code: 3cli
was solved by
D.-S.Lee,
P.Nioche,
C.S.Raman,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 88.39 / 1.80 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 63.639, 105.192, 162.295, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17 / 19.8 |
Iron Binding Sites:
The binding sites of Iron atom in the Crystal Structure of Arabidopsis Thaliana Allene Oxide Synthase (Aos, Cytochrome P450 74A, CYP74A) at 1.80 A Resolution
(pdb code 3cli). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 2 binding sites of Iron where determined in the Crystal Structure of Arabidopsis Thaliana Allene Oxide Synthase (Aos, Cytochrome P450 74A, CYP74A) at 1.80 A Resolution, PDB code: 3cli:
Jump to Iron binding site number: 1; 2;
In total 2 binding sites of Iron where determined in the Crystal Structure of Arabidopsis Thaliana Allene Oxide Synthase (Aos, Cytochrome P450 74A, CYP74A) at 1.80 A Resolution, PDB code: 3cli:
Jump to Iron binding site number: 1; 2;
Iron binding site 1 out of 2 in 3cli
Go back to
Iron binding site 1 out
of 2 in the Crystal Structure of Arabidopsis Thaliana Allene Oxide Synthase (Aos, Cytochrome P450 74A, CYP74A) at 1.80 A Resolution
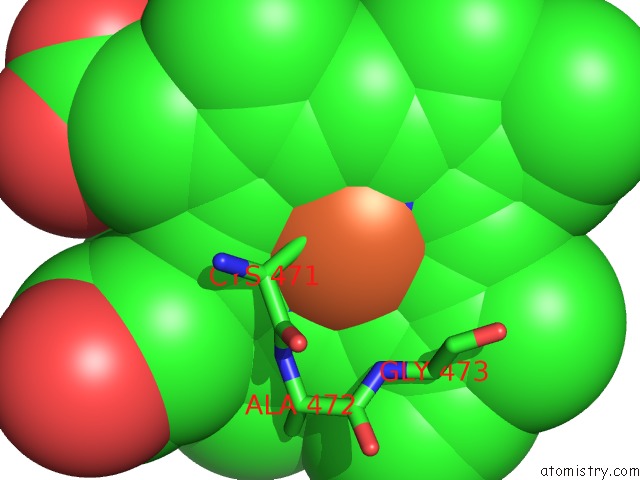
Mono view
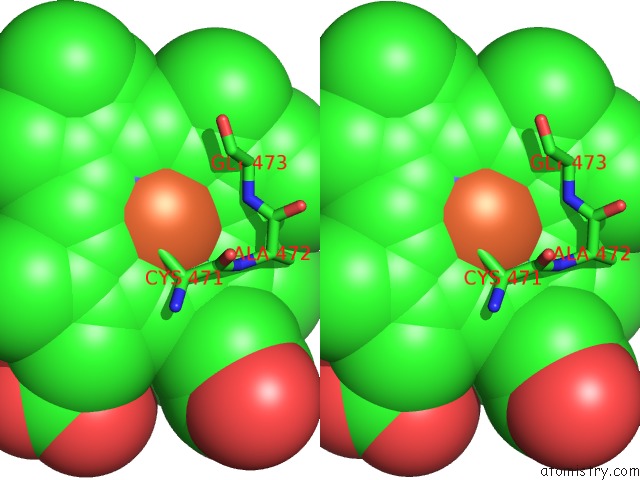
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Crystal Structure of Arabidopsis Thaliana Allene Oxide Synthase (Aos, Cytochrome P450 74A, CYP74A) at 1.80 A Resolution within 5.0Å range:
|
Iron binding site 2 out of 2 in 3cli
Go back to
Iron binding site 2 out
of 2 in the Crystal Structure of Arabidopsis Thaliana Allene Oxide Synthase (Aos, Cytochrome P450 74A, CYP74A) at 1.80 A Resolution
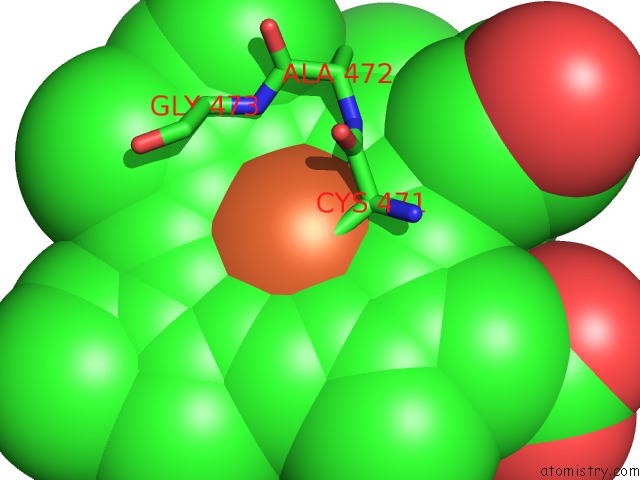
Mono view
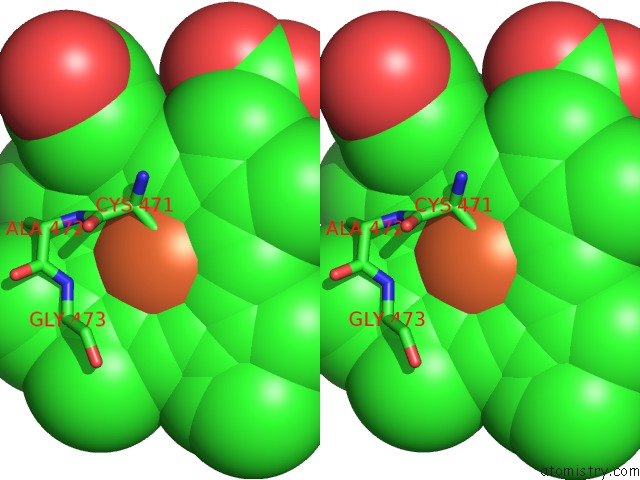
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Crystal Structure of Arabidopsis Thaliana Allene Oxide Synthase (Aos, Cytochrome P450 74A, CYP74A) at 1.80 A Resolution within 5.0Å range:
|
Reference:
D.-S.Lee,
P.Nioche,
M.Hamberg,
C.S.Raman.
Structural Insights Into the Evolutionary Paths of Oxylipin Biosynthetic Enzymes Nature V. 455 363 2008.
ISSN: ISSN 0028-0836
PubMed: 18716621
DOI: 10.1038/NATURE07307
Page generated: Sun Aug 4 08:24:15 2024
ISSN: ISSN 0028-0836
PubMed: 18716621
DOI: 10.1038/NATURE07307
Last articles
F in 7LOMF in 7LOD
F in 7LOC
F in 7LO6
F in 7LOA
F in 7LOB
F in 7LK1
F in 7LL4
F in 7LKL
F in 7LIU