Iron »
PDB 3etr-3fou »
3fgs »
Iron in PDB 3fgs: Crystal Structure of G65R/K206E Double Mutant of the N-Lobe Human Transferrin
Protein crystallography data
The structure of Crystal Structure of G65R/K206E Double Mutant of the N-Lobe Human Transferrin, PDB code: 3fgs
was solved by
P.J.Halbrooks,
A.B.Mason,
S.J.Everse,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 19.49 / 1.80 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 43.347, 57.056, 133.737, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 21.6 / 24.6 |
Iron Binding Sites:
The binding sites of Iron atom in the Crystal Structure of G65R/K206E Double Mutant of the N-Lobe Human Transferrin
(pdb code 3fgs). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total only one binding site of Iron was determined in the Crystal Structure of G65R/K206E Double Mutant of the N-Lobe Human Transferrin, PDB code: 3fgs:
In total only one binding site of Iron was determined in the Crystal Structure of G65R/K206E Double Mutant of the N-Lobe Human Transferrin, PDB code: 3fgs:
Iron binding site 1 out of 1 in 3fgs
Go back to
Iron binding site 1 out
of 1 in the Crystal Structure of G65R/K206E Double Mutant of the N-Lobe Human Transferrin
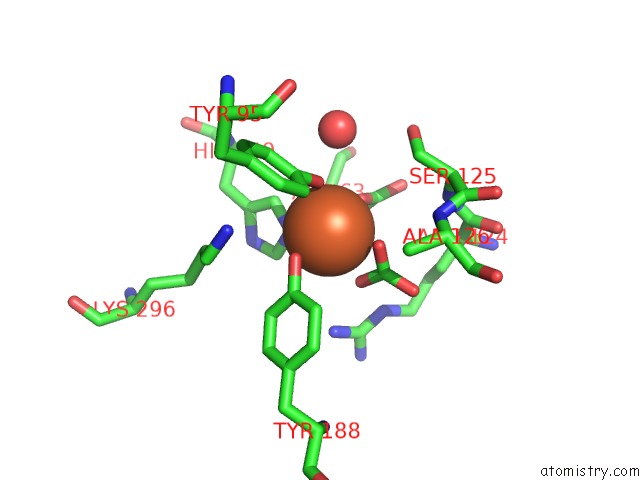
Mono view
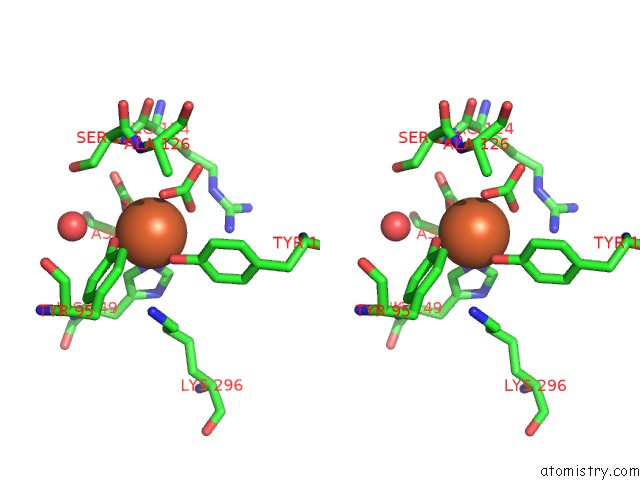
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Crystal Structure of G65R/K206E Double Mutant of the N-Lobe Human Transferrin within 5.0Å range:
|
Reference:
A.B.Mason,
P.J.Halbrooks,
N.G.James,
S.L.Byrne,
J.K.Grady,
N.D.Chasteen,
C.E.Bobst,
I.A.Kaltashov,
V.C.Smith,
R.T.Macgillivray,
S.J.Everse.
Structural and Functional Consequences of the Substitution of Glycine 65 with Arginine in the N-Lobe of Human Transferrin. Biochemistry V. 48 1945 2009.
ISSN: ISSN 0006-2960
PubMed: 19219998
DOI: 10.1021/BI802254X
Page generated: Sun Aug 4 10:04:57 2024
ISSN: ISSN 0006-2960
PubMed: 19219998
DOI: 10.1021/BI802254X
Last articles
Cl in 3TFPCl in 3TFC
Cl in 3TCT
Cl in 3TF6
Cl in 3TDX
Cl in 3TEE
Cl in 3TD5
Cl in 3TDJ
Cl in 3TDQ
Cl in 3TCS