Iron »
PDB 3etr-3fou »
3fmr »
Iron in PDB 3fmr: Crystal Structure of An Encephalitozoon Cuniculi Methionine Aminopeptidase Type 2 with Angiogenesis Inhibitor TNP470 Bound
Enzymatic activity of Crystal Structure of An Encephalitozoon Cuniculi Methionine Aminopeptidase Type 2 with Angiogenesis Inhibitor TNP470 Bound
All present enzymatic activity of Crystal Structure of An Encephalitozoon Cuniculi Methionine Aminopeptidase Type 2 with Angiogenesis Inhibitor TNP470 Bound:
3.4.11.18;
3.4.11.18;
Protein crystallography data
The structure of Crystal Structure of An Encephalitozoon Cuniculi Methionine Aminopeptidase Type 2 with Angiogenesis Inhibitor TNP470 Bound, PDB code: 3fmr
was solved by
J.J.Alvarado,
M.Russell,
A.Zhang,
J.Adams,
R.Toro,
S.K.Burley,
L.M.Weiss,
S.C.Almo,
New York Sgx Research Center For Structural Genomics(Nysgxrc),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 2.89 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 61.977, 94.067, 66.407, 90.00, 99.33, 90.00 |
| R / Rfree (%) | 17.8 / 26.6 |
Iron Binding Sites:
The binding sites of Iron atom in the Crystal Structure of An Encephalitozoon Cuniculi Methionine Aminopeptidase Type 2 with Angiogenesis Inhibitor TNP470 Bound
(pdb code 3fmr). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 4 binding sites of Iron where determined in the Crystal Structure of An Encephalitozoon Cuniculi Methionine Aminopeptidase Type 2 with Angiogenesis Inhibitor TNP470 Bound, PDB code: 3fmr:
Jump to Iron binding site number: 1; 2; 3; 4;
In total 4 binding sites of Iron where determined in the Crystal Structure of An Encephalitozoon Cuniculi Methionine Aminopeptidase Type 2 with Angiogenesis Inhibitor TNP470 Bound, PDB code: 3fmr:
Jump to Iron binding site number: 1; 2; 3; 4;
Iron binding site 1 out of 4 in 3fmr
Go back to
Iron binding site 1 out
of 4 in the Crystal Structure of An Encephalitozoon Cuniculi Methionine Aminopeptidase Type 2 with Angiogenesis Inhibitor TNP470 Bound
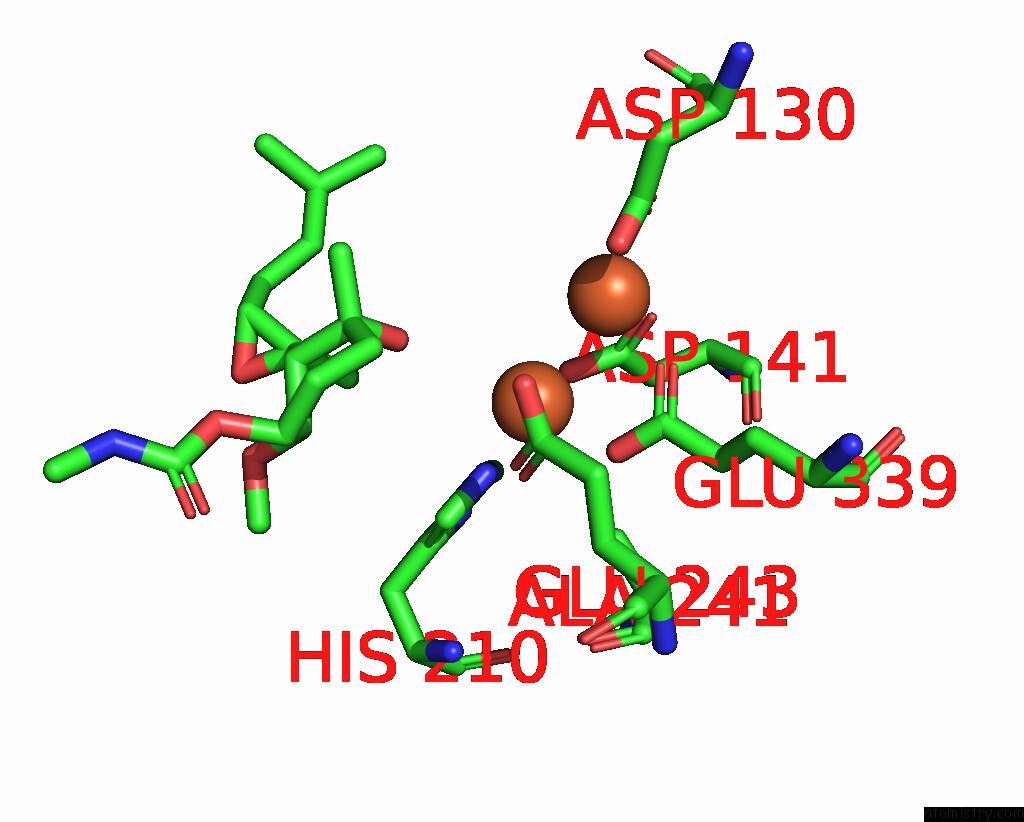
Mono view
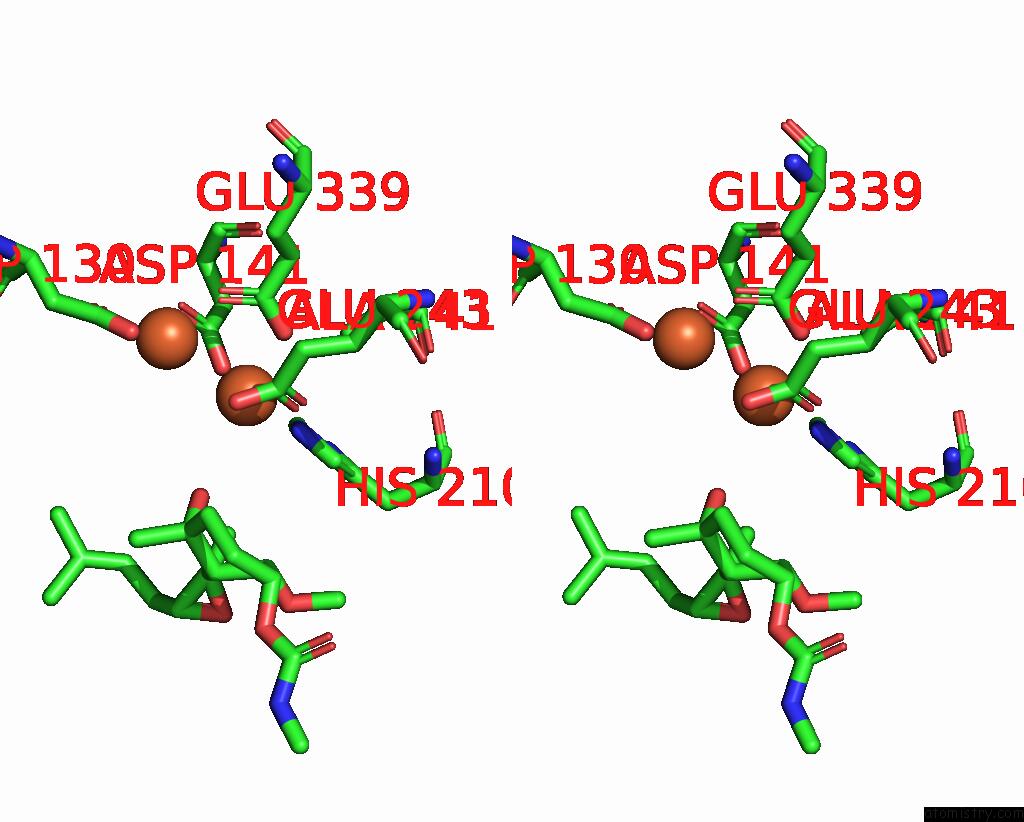
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Crystal Structure of An Encephalitozoon Cuniculi Methionine Aminopeptidase Type 2 with Angiogenesis Inhibitor TNP470 Bound within 5.0Å range:
|
Iron binding site 2 out of 4 in 3fmr
Go back to
Iron binding site 2 out
of 4 in the Crystal Structure of An Encephalitozoon Cuniculi Methionine Aminopeptidase Type 2 with Angiogenesis Inhibitor TNP470 Bound
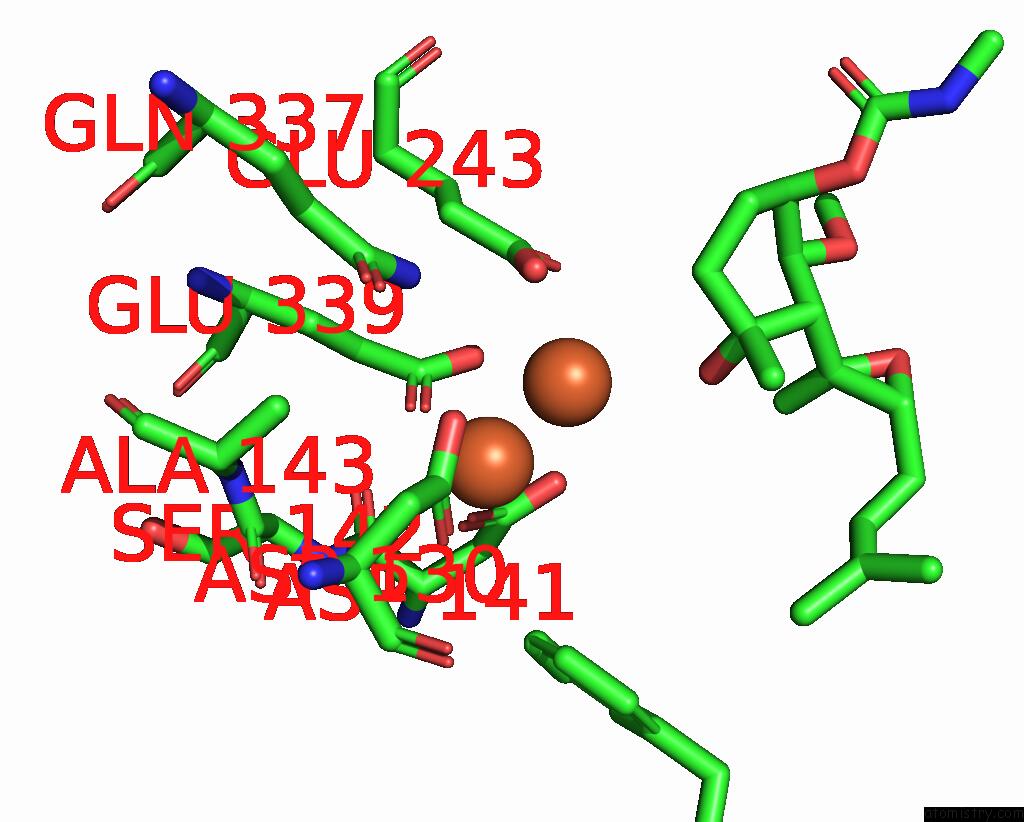
Mono view
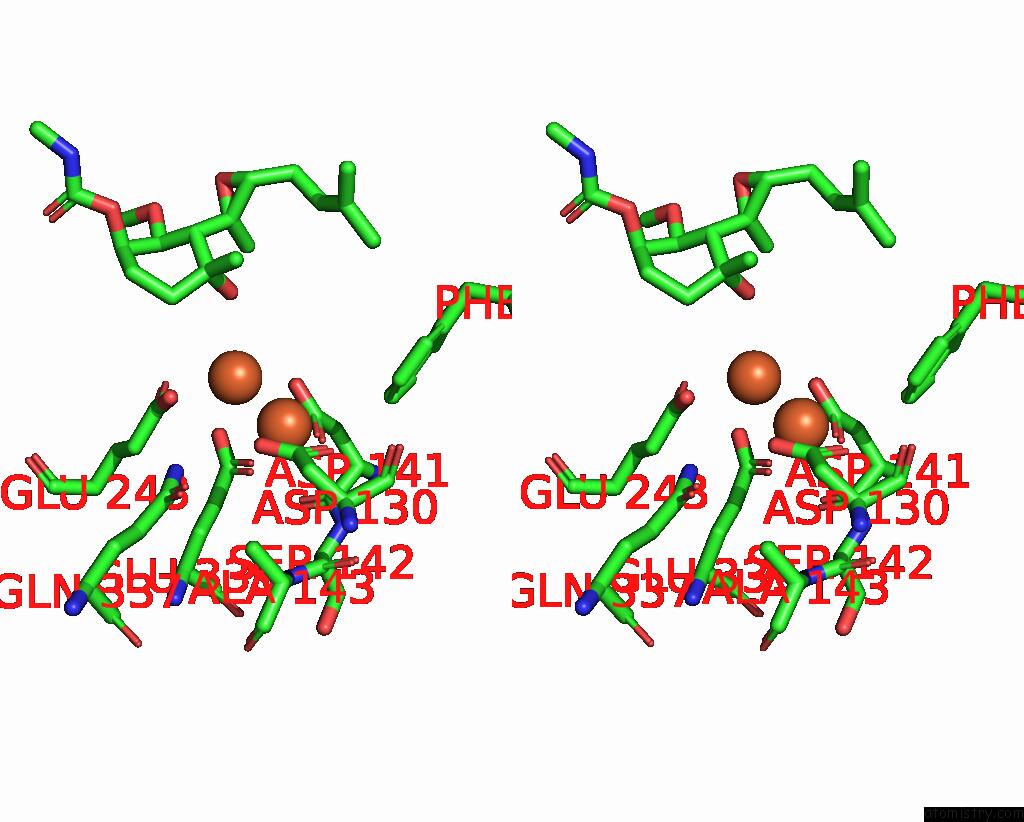
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Crystal Structure of An Encephalitozoon Cuniculi Methionine Aminopeptidase Type 2 with Angiogenesis Inhibitor TNP470 Bound within 5.0Å range:
|
Iron binding site 3 out of 4 in 3fmr
Go back to
Iron binding site 3 out
of 4 in the Crystal Structure of An Encephalitozoon Cuniculi Methionine Aminopeptidase Type 2 with Angiogenesis Inhibitor TNP470 Bound
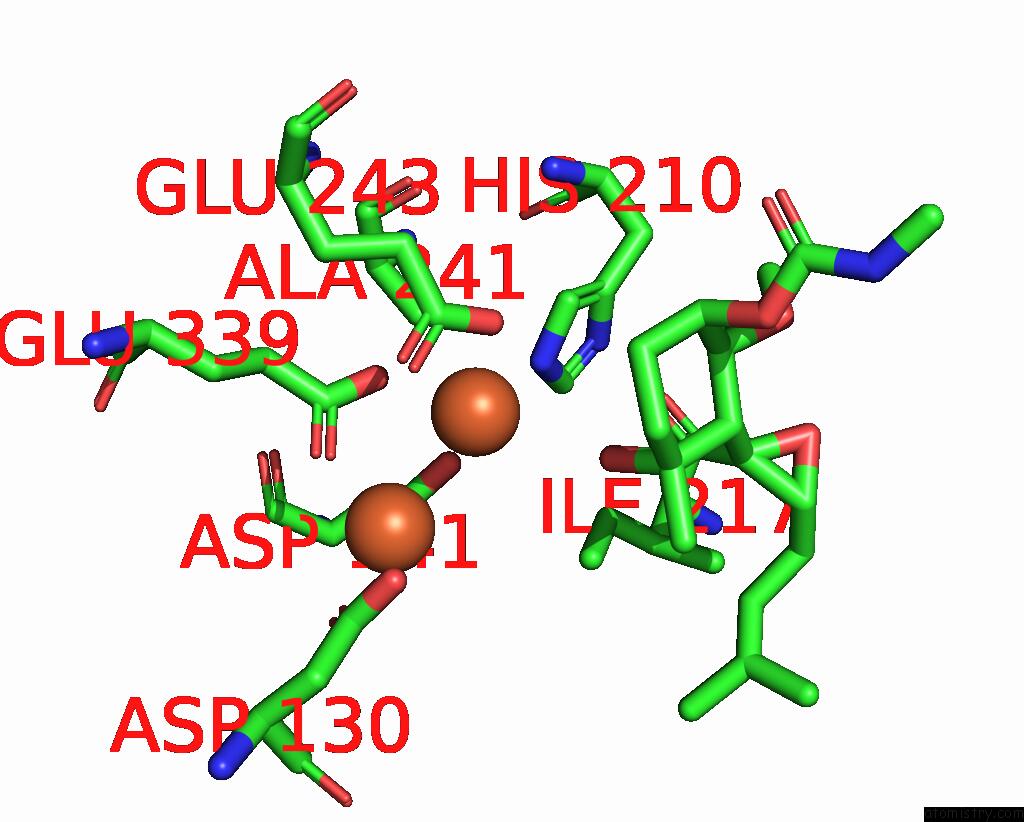
Mono view
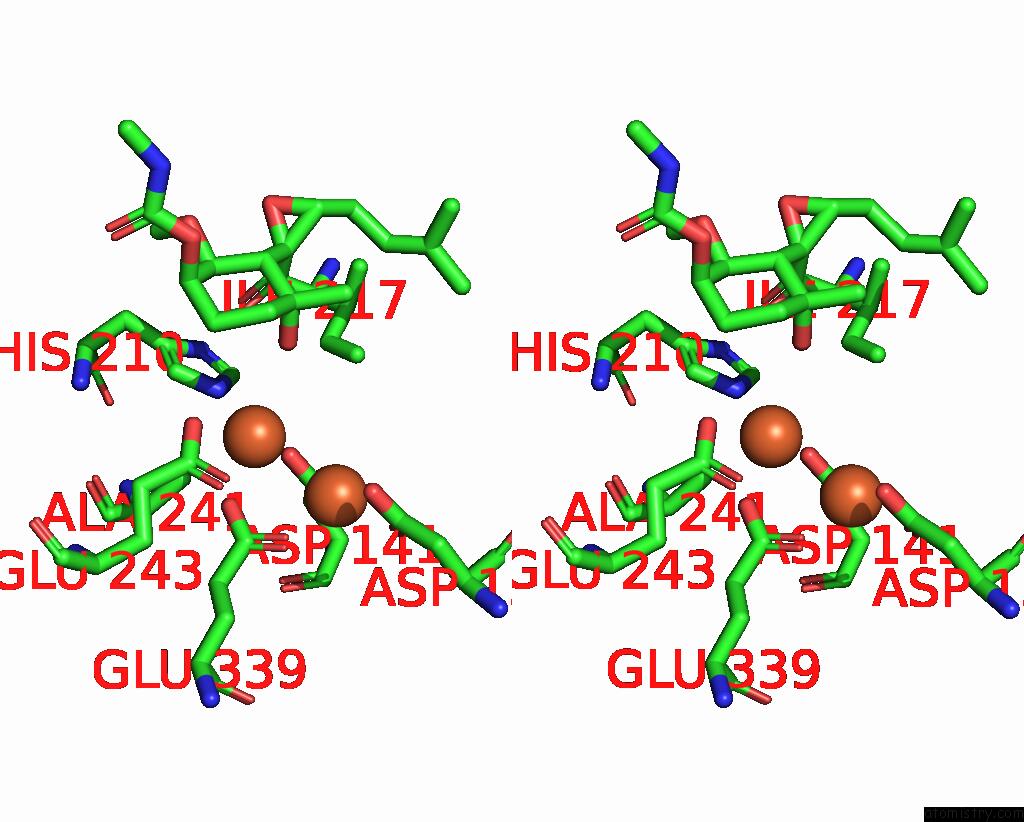
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 3 of Crystal Structure of An Encephalitozoon Cuniculi Methionine Aminopeptidase Type 2 with Angiogenesis Inhibitor TNP470 Bound within 5.0Å range:
|
Iron binding site 4 out of 4 in 3fmr
Go back to
Iron binding site 4 out
of 4 in the Crystal Structure of An Encephalitozoon Cuniculi Methionine Aminopeptidase Type 2 with Angiogenesis Inhibitor TNP470 Bound
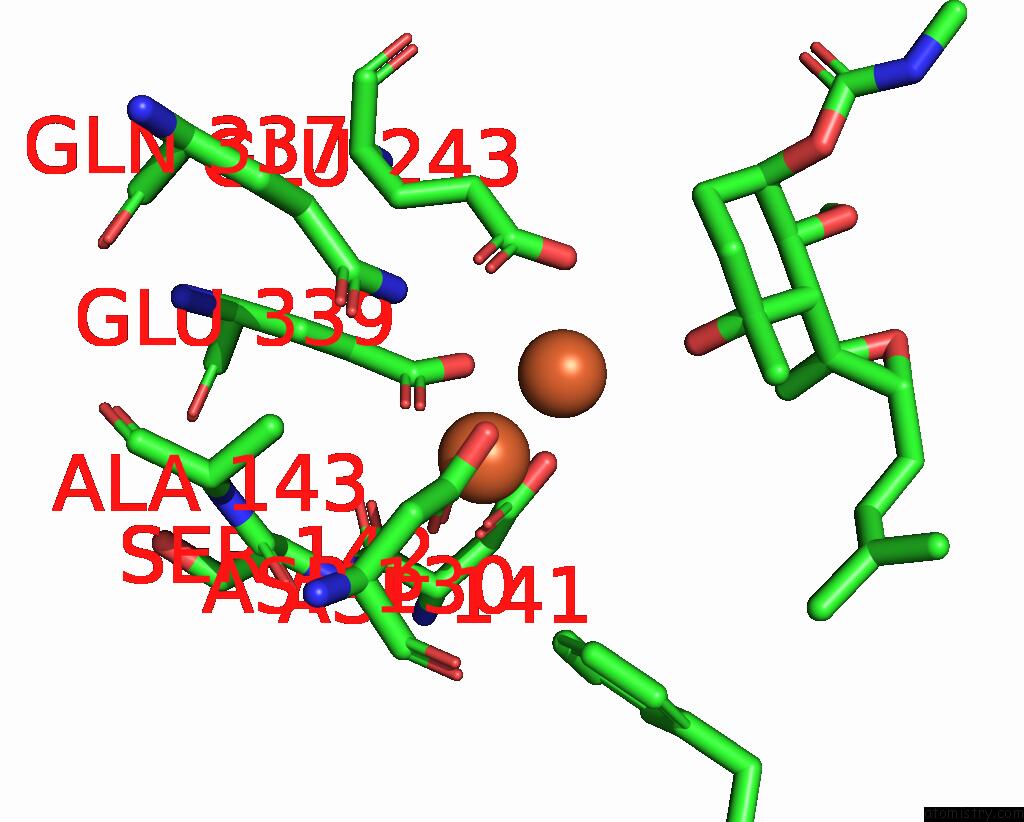
Mono view
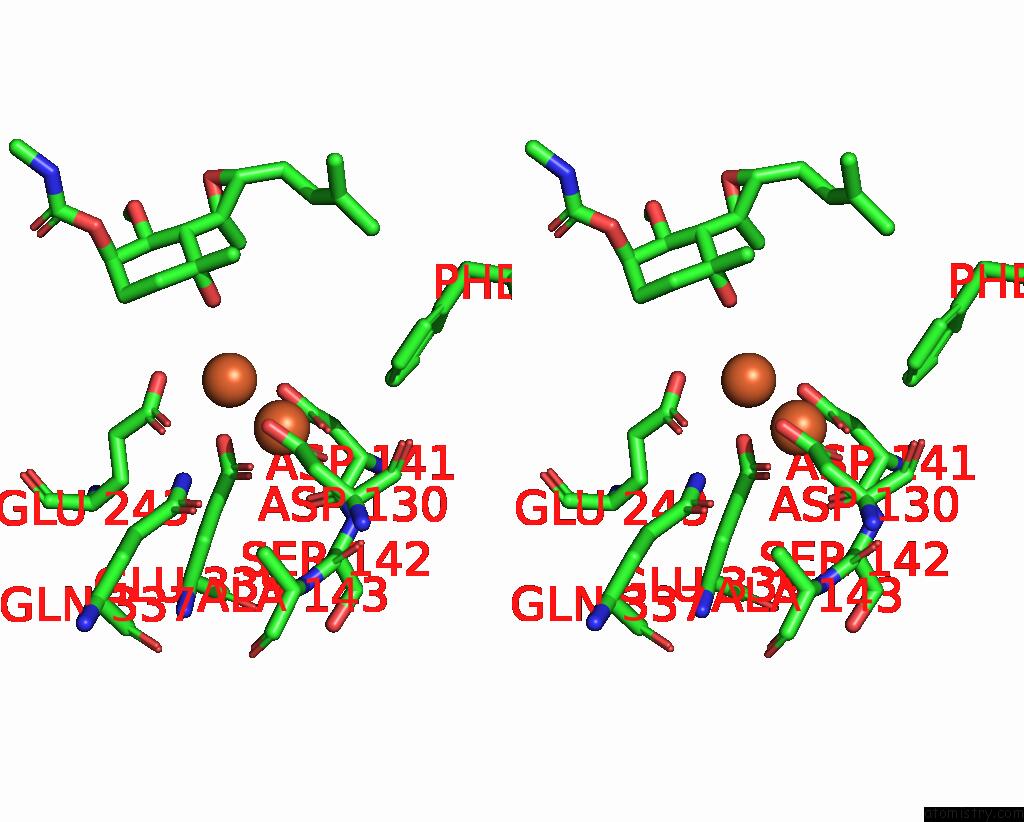
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 4 of Crystal Structure of An Encephalitozoon Cuniculi Methionine Aminopeptidase Type 2 with Angiogenesis Inhibitor TNP470 Bound within 5.0Å range:
|
Reference:
J.J.Alvarado,
A.Nemkal,
J.M.Sauder,
M.Russell,
D.E.Akiyoshi,
W.Shi,
S.C.Almo,
L.M.Weiss.
Structure of A Microsporidian Methionine Aminopeptidase Type 2 Complexed with Fumagillin and Tnp-470. Mol.Biochem.Parasitol. V. 168 158 2009.
ISSN: ISSN 0166-6851
PubMed: 19660503
DOI: 10.1016/J.MOLBIOPARA.2009.07.008
Page generated: Sun Aug 4 10:07:37 2024
ISSN: ISSN 0166-6851
PubMed: 19660503
DOI: 10.1016/J.MOLBIOPARA.2009.07.008
Last articles
Cl in 5VMZCl in 5VMY
Cl in 5VMX
Cl in 5VMW
Cl in 5VMQ
Cl in 5VMV
Cl in 5VMU
Cl in 5VL2
Cl in 5VKW
Cl in 5VM0