Iron »
PDB 3nmk-3nwv »
3nml »
Iron in PDB 3nml: Sperm Whale Myoglobin Mutant H64W Carbonmonoxy-Form
Protein crystallography data
The structure of Sperm Whale Myoglobin Mutant H64W Carbonmonoxy-Form, PDB code: 3nml
was solved by
I.Birukou,
J.Soman,
J.S.Olson,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 22.71 / 1.68 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 38.930, 46.960, 89.500, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 16.6 / 19.6 |
Iron Binding Sites:
The binding sites of Iron atom in the Sperm Whale Myoglobin Mutant H64W Carbonmonoxy-Form
(pdb code 3nml). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total only one binding site of Iron was determined in the Sperm Whale Myoglobin Mutant H64W Carbonmonoxy-Form, PDB code: 3nml:
In total only one binding site of Iron was determined in the Sperm Whale Myoglobin Mutant H64W Carbonmonoxy-Form, PDB code: 3nml:
Iron binding site 1 out of 1 in 3nml
Go back to
Iron binding site 1 out
of 1 in the Sperm Whale Myoglobin Mutant H64W Carbonmonoxy-Form
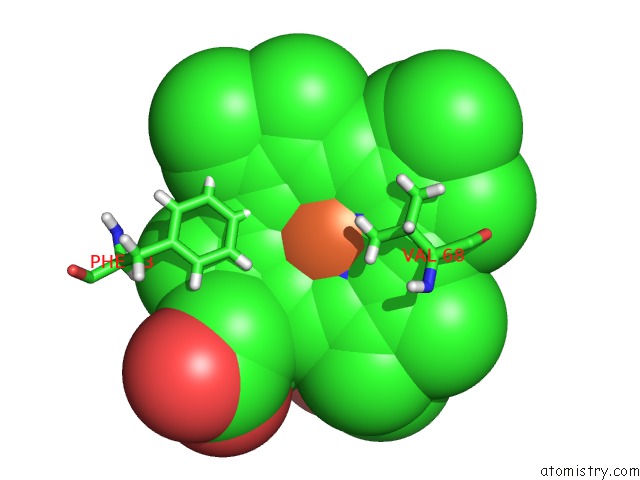
Mono view
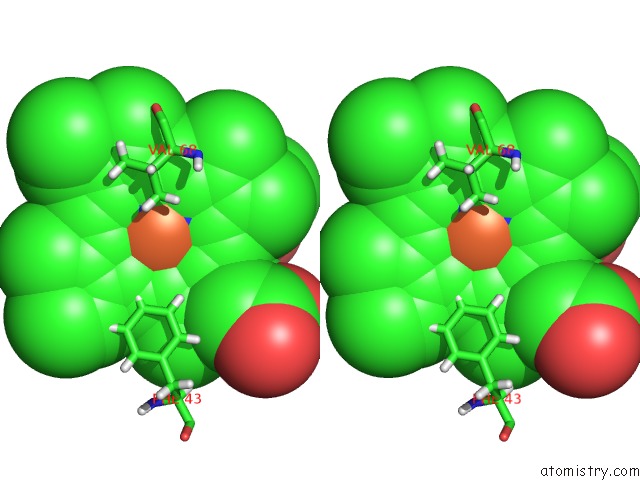
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Sperm Whale Myoglobin Mutant H64W Carbonmonoxy-Form within 5.0Å range:
|
Reference:
I.Birukou,
J.Soman,
J.S.Olson.
Blocking the Gate to Ligand Entry in Human Hemoglobin. J.Biol.Chem. V. 286 10515 2011.
ISSN: ISSN 0021-9258
PubMed: 21193395
DOI: 10.1074/JBC.M110.176271
Page generated: Tue Aug 5 04:50:03 2025
ISSN: ISSN 0021-9258
PubMed: 21193395
DOI: 10.1074/JBC.M110.176271
Last articles
Fe in 3VYTFe in 3VYU
Fe in 3W1W
Fe in 3W08
Fe in 3VYS
Fe in 3VYR
Fe in 3VV9
Fe in 3VVA
Fe in 3VYM
Fe in 3VXJ