Iron »
PDB 3pcq-3puq »
3ph2 »
Iron in PDB 3ph2: Structure of the Imidazole-Adduct of the Phormidium Laminosum Cytochrome C6 Q51V Variant
Protein crystallography data
The structure of Structure of the Imidazole-Adduct of the Phormidium Laminosum Cytochrome C6 Q51V Variant, PDB code: 3ph2
was solved by
J.A.R.Worrall,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 25.03 / 1.40 |
| Space group | P 65 |
| Cell size a, b, c (Å), α, β, γ (°) | 45.616, 45.616, 64.707, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 18.6 / 22.2 |
Iron Binding Sites:
The binding sites of Iron atom in the Structure of the Imidazole-Adduct of the Phormidium Laminosum Cytochrome C6 Q51V Variant
(pdb code 3ph2). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total only one binding site of Iron was determined in the Structure of the Imidazole-Adduct of the Phormidium Laminosum Cytochrome C6 Q51V Variant, PDB code: 3ph2:
In total only one binding site of Iron was determined in the Structure of the Imidazole-Adduct of the Phormidium Laminosum Cytochrome C6 Q51V Variant, PDB code: 3ph2:
Iron binding site 1 out of 1 in 3ph2
Go back to
Iron binding site 1 out
of 1 in the Structure of the Imidazole-Adduct of the Phormidium Laminosum Cytochrome C6 Q51V Variant
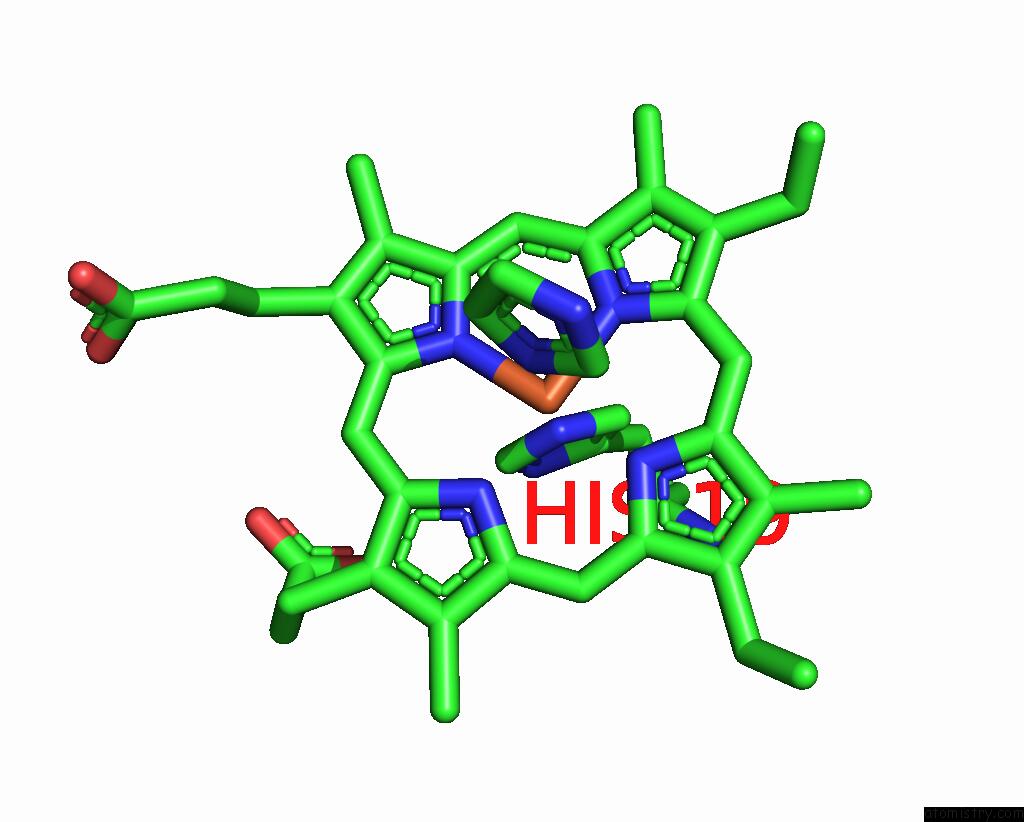
Mono view
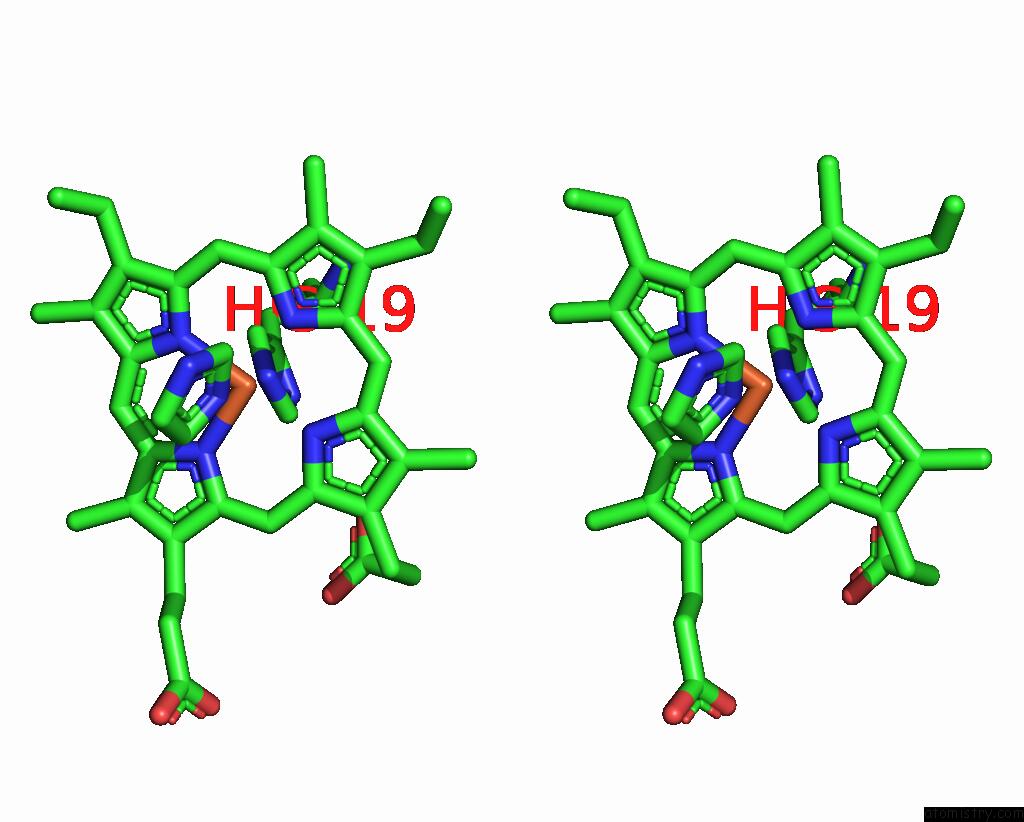
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Structure of the Imidazole-Adduct of the Phormidium Laminosum Cytochrome C6 Q51V Variant within 5.0Å range:
|
Reference:
B.S.Rajagopal,
M.T.Wilson,
D.S.Bendall,
C.J.Howe,
J.A.Worrall.
Structural and Kinetic Studies of Imidazole Binding to Two Members of the Cytochrome C (6) Family Reveal An Important Role For A Conserved Heme Pocket Residue. J.Biol.Inorg.Chem. V. 16 577 2011.
ISSN: ISSN 0949-8257
PubMed: 21267610
DOI: 10.1007/S00775-011-0758-Y
Page generated: Tue Aug 5 05:42:58 2025
ISSN: ISSN 0949-8257
PubMed: 21267610
DOI: 10.1007/S00775-011-0758-Y
Last articles
Fe in 3WCPFe in 3WCQ
Fe in 3WC8
Fe in 3WAH
Fe in 3WAQ
Fe in 3W9C
Fe in 3W5U
Fe in 3W8O
Fe in 3WAE
Fe in 3W54