Iron »
PDB 3tgu-3u3e »
3tm3 »
Iron in PDB 3tm3: Wild-Type Hemoglobin From Vitreoscilla Stercoraria
Protein crystallography data
The structure of Wild-Type Hemoglobin From Vitreoscilla Stercoraria, PDB code: 3tm3
was solved by
S.Ratakonda,
A.Anand,
K.Dikshit,
B.C.Stark,
A.J.Howard,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.93 / 1.75 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 75.156, 99.013, 41.015, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17 / 20.4 |
Iron Binding Sites:
The binding sites of Iron atom in the Wild-Type Hemoglobin From Vitreoscilla Stercoraria
(pdb code 3tm3). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total only one binding site of Iron was determined in the Wild-Type Hemoglobin From Vitreoscilla Stercoraria, PDB code: 3tm3:
In total only one binding site of Iron was determined in the Wild-Type Hemoglobin From Vitreoscilla Stercoraria, PDB code: 3tm3:
Iron binding site 1 out of 1 in 3tm3
Go back to
Iron binding site 1 out
of 1 in the Wild-Type Hemoglobin From Vitreoscilla Stercoraria
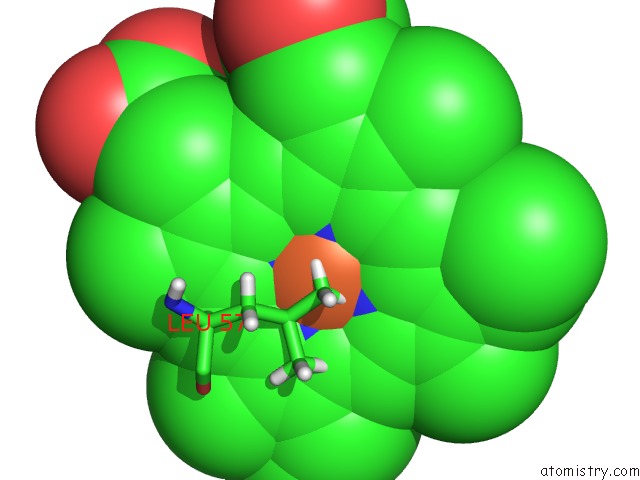
Mono view
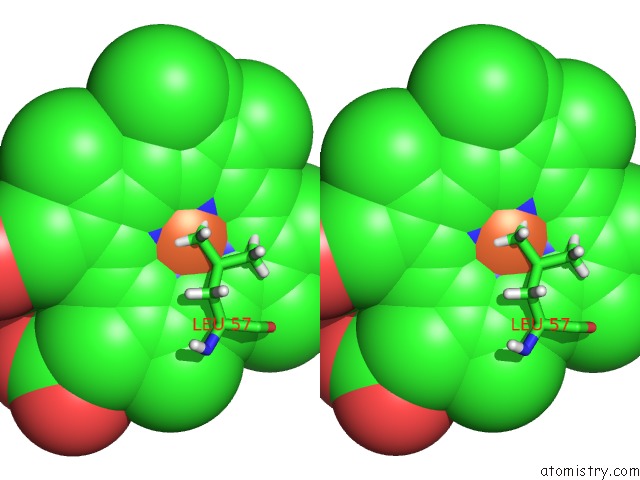
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Wild-Type Hemoglobin From Vitreoscilla Stercoraria within 5.0Å range:
|
Reference:
S.Ratakonda,
A.Anand,
K.Dikshit,
B.C.Stark,
A.J.Howard.
Crystallographic Structure Determination of B10 Mutants of Vitreoscilla Hemoglobin: Role of TYR29 (B10) in the Structure of the Ligand-Binding Site. Acta Crystallogr.,Sect.F V. 69 215 2013.
ISSN: ESSN 1744-3091
PubMed: 23519792
DOI: 10.1107/S1744309112044818
Page generated: Sun Aug 4 20:29:30 2024
ISSN: ESSN 1744-3091
PubMed: 23519792
DOI: 10.1107/S1744309112044818
Last articles
Cl in 5W91Cl in 5W8R
Cl in 5W8C
Cl in 5W8E
Cl in 5W80
Cl in 5W7U
Cl in 5W7K
Cl in 5W6T
Cl in 5W71
Cl in 5W73