Iron »
PDB 4bmq-4ccp »
4c50 »
Iron in PDB 4c50: Crystal Structure of the Catalase-Peroxidase (Katg) D137S Mutant From Mycobacterium Tuberculosis
Enzymatic activity of Crystal Structure of the Catalase-Peroxidase (Katg) D137S Mutant From Mycobacterium Tuberculosis
All present enzymatic activity of Crystal Structure of the Catalase-Peroxidase (Katg) D137S Mutant From Mycobacterium Tuberculosis:
1.11.1.21;
1.11.1.21;
Protein crystallography data
The structure of Crystal Structure of the Catalase-Peroxidase (Katg) D137S Mutant From Mycobacterium Tuberculosis, PDB code: 4c50
was solved by
H.-P.Hersleth,
X.Zhao,
R.S.Magliozzo,
K.K.Andersson,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 75.41 / 2.50 |
| Space group | P 42 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 150.590, 150.590, 157.350, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 26.363 / 31.113 |
Iron Binding Sites:
The binding sites of Iron atom in the Crystal Structure of the Catalase-Peroxidase (Katg) D137S Mutant From Mycobacterium Tuberculosis
(pdb code 4c50). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 2 binding sites of Iron where determined in the Crystal Structure of the Catalase-Peroxidase (Katg) D137S Mutant From Mycobacterium Tuberculosis, PDB code: 4c50:
Jump to Iron binding site number: 1; 2;
In total 2 binding sites of Iron where determined in the Crystal Structure of the Catalase-Peroxidase (Katg) D137S Mutant From Mycobacterium Tuberculosis, PDB code: 4c50:
Jump to Iron binding site number: 1; 2;
Iron binding site 1 out of 2 in 4c50
Go back to
Iron binding site 1 out
of 2 in the Crystal Structure of the Catalase-Peroxidase (Katg) D137S Mutant From Mycobacterium Tuberculosis
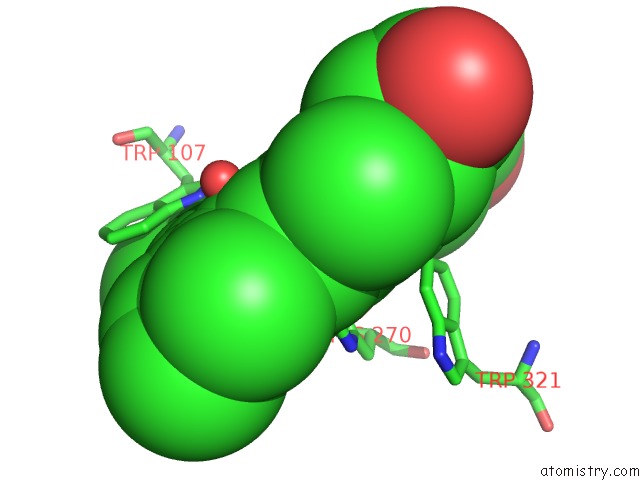
Mono view
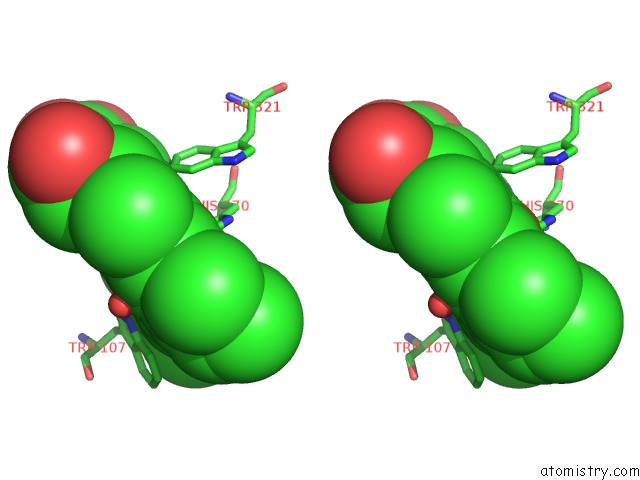
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Crystal Structure of the Catalase-Peroxidase (Katg) D137S Mutant From Mycobacterium Tuberculosis within 5.0Å range:
|
Iron binding site 2 out of 2 in 4c50
Go back to
Iron binding site 2 out
of 2 in the Crystal Structure of the Catalase-Peroxidase (Katg) D137S Mutant From Mycobacterium Tuberculosis
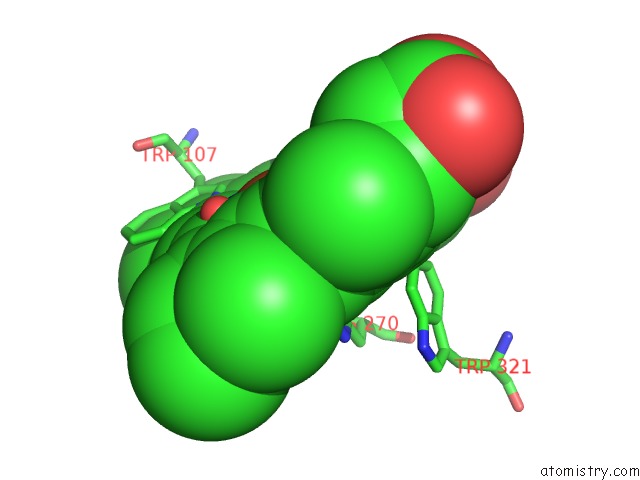
Mono view
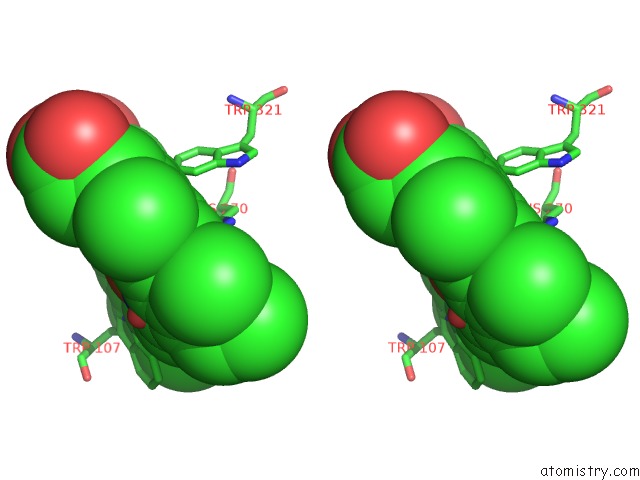
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Crystal Structure of the Catalase-Peroxidase (Katg) D137S Mutant From Mycobacterium Tuberculosis within 5.0Å range:
|
Reference:
X.Zhao,
H.P.Hersleth,
J.Zhu,
K.K.Andersson,
R.S.Magliozzo.
Access Channel Residues SER315 and ASP137 in Mycobacterium Tuberculosis Catalase-Peroxidase (Katg) Control Peroxidatic Activation of the Pro-Drug Isoniazid. Chem.Commun.(Camb.) V. 49 11650 2013.
ISSN: ISSN 1359-7345
PubMed: 24185282
DOI: 10.1039/C3CC47022A
Page generated: Tue Aug 5 09:19:47 2025
ISSN: ISSN 1359-7345
PubMed: 24185282
DOI: 10.1039/C3CC47022A
Last articles
Fe in 5LBHFe in 5LC8
Fe in 5L8R
Fe in 5L94
Fe in 5L92
Fe in 5L2R
Fe in 5L90
Fe in 5L91
Fe in 5L86
Fe in 5L8D