Iron »
PDB 4g39-4gl5 »
4g7u »
Iron in PDB 4g7u: Rat Heme Oxygenase-1 in Complex with Heme and Co with 16 Hr Illumination: Laser Off
Enzymatic activity of Rat Heme Oxygenase-1 in Complex with Heme and Co with 16 Hr Illumination: Laser Off
All present enzymatic activity of Rat Heme Oxygenase-1 in Complex with Heme and Co with 16 Hr Illumination: Laser Off:
1.14.99.3;
1.14.99.3;
Protein crystallography data
The structure of Rat Heme Oxygenase-1 in Complex with Heme and Co with 16 Hr Illumination: Laser Off, PDB code: 4g7u
was solved by
M.Sugishima,
K.Moffat,
M.Noguchi,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 32.91 / 1.90 |
| Space group | P 32 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 65.819, 65.819, 119.879, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 16.2 / 20.5 |
Iron Binding Sites:
The binding sites of Iron atom in the Rat Heme Oxygenase-1 in Complex with Heme and Co with 16 Hr Illumination: Laser Off
(pdb code 4g7u). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total only one binding site of Iron was determined in the Rat Heme Oxygenase-1 in Complex with Heme and Co with 16 Hr Illumination: Laser Off, PDB code: 4g7u:
In total only one binding site of Iron was determined in the Rat Heme Oxygenase-1 in Complex with Heme and Co with 16 Hr Illumination: Laser Off, PDB code: 4g7u:
Iron binding site 1 out of 1 in 4g7u
Go back to
Iron binding site 1 out
of 1 in the Rat Heme Oxygenase-1 in Complex with Heme and Co with 16 Hr Illumination: Laser Off
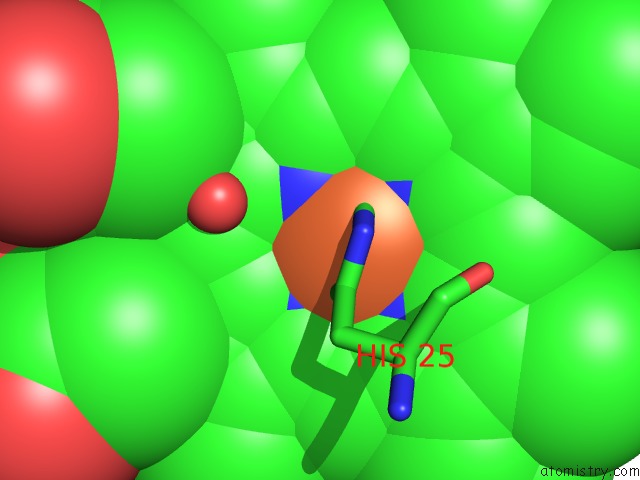
Mono view
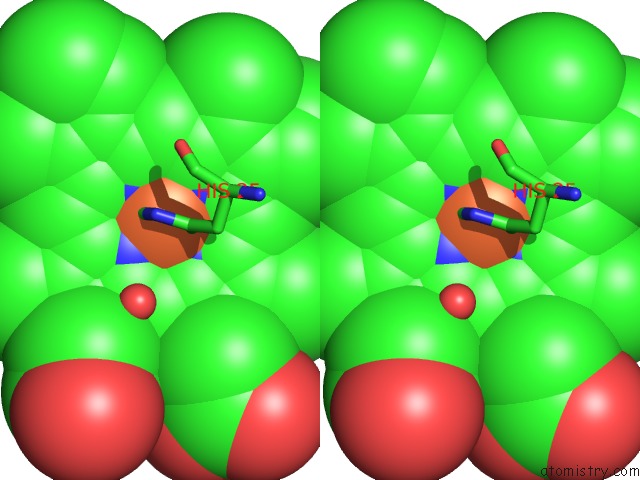
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Rat Heme Oxygenase-1 in Complex with Heme and Co with 16 Hr Illumination: Laser Off within 5.0Å range:
|
Reference:
M.Sugishima,
K.Moffat,
M.Noguchi.
Discrimination Between Co and O2 in Heme Oxygenase: Comparison of Static Structures and Dynamic Conformation Changes Following Co Photolysis Biochemistry 2012.
ISSN: ISSN 0006-2960
PubMed: 23043644
DOI: 10.1021/BI301175X
Page generated: Tue Aug 5 10:35:52 2025
ISSN: ISSN 0006-2960
PubMed: 23043644
DOI: 10.1021/BI301175X
Last articles
Fe in 5TH5Fe in 5T5M
Fe in 5TIA
Fe in 5TI9
Fe in 5TGS
Fe in 5TFU
Fe in 5TG0
Fe in 5TFT
Fe in 5TE8
Fe in 5TDV