Iron »
PDB 4lm8-4m6y »
4lxj »
Iron in PDB 4lxj: Saccharomyces Cerevisiae Lanosterol 14-Alpha Demethylase with Lanosterol Bound
Enzymatic activity of Saccharomyces Cerevisiae Lanosterol 14-Alpha Demethylase with Lanosterol Bound
All present enzymatic activity of Saccharomyces Cerevisiae Lanosterol 14-Alpha Demethylase with Lanosterol Bound:
1.14.13.70;
1.14.13.70;
Protein crystallography data
The structure of Saccharomyces Cerevisiae Lanosterol 14-Alpha Demethylase with Lanosterol Bound, PDB code: 4lxj
was solved by
B.C.Monk,
T.M.Tomasiak,
M.V.Keniya,
F.U.Huschmann,
J.D.A.Tyndall,
J.D.O'connell Iii,
R.D.Cannon,
J.Mcdonald,
A.Rodriguez,
J.Finer-Moore,
R.M.Stroud,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.33 / 1.90 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 78.213, 67.005, 80.378, 90.00, 99.54, 90.00 |
| R / Rfree (%) | 19.5 / 22.7 |
Iron Binding Sites:
The binding sites of Iron atom in the Saccharomyces Cerevisiae Lanosterol 14-Alpha Demethylase with Lanosterol Bound
(pdb code 4lxj). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total only one binding site of Iron was determined in the Saccharomyces Cerevisiae Lanosterol 14-Alpha Demethylase with Lanosterol Bound, PDB code: 4lxj:
In total only one binding site of Iron was determined in the Saccharomyces Cerevisiae Lanosterol 14-Alpha Demethylase with Lanosterol Bound, PDB code: 4lxj:
Iron binding site 1 out of 1 in 4lxj
Go back to
Iron binding site 1 out
of 1 in the Saccharomyces Cerevisiae Lanosterol 14-Alpha Demethylase with Lanosterol Bound
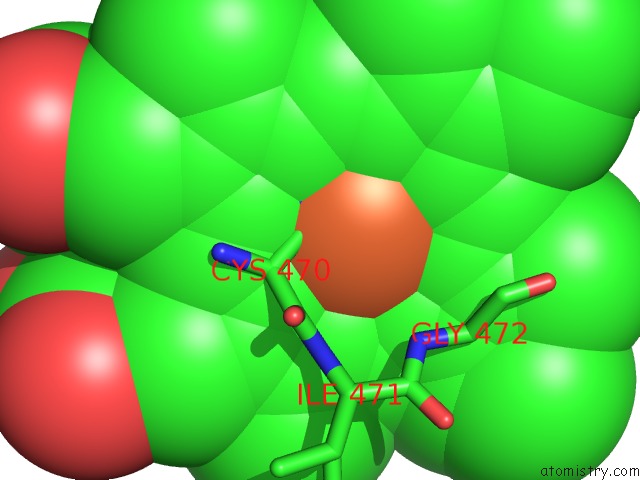
Mono view
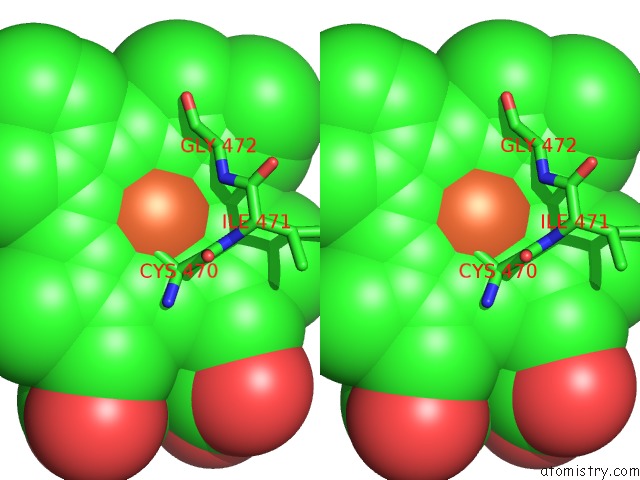
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Saccharomyces Cerevisiae Lanosterol 14-Alpha Demethylase with Lanosterol Bound within 5.0Å range:
|
Reference:
B.C.Monk,
T.M.Tomasiak,
M.V.Keniya,
F.U.Huschmann,
J.D.Tyndall,
J.D.O'connell,
R.D.Cannon,
J.G.Mcdonald,
A.Rodriguez,
J.S.Finer-Moore,
R.M.Stroud.
Architecture of A Single Membrane Spanning Cytochrome P450 Suggests Constraints That Orient the Catalytic Domain Relative to A Bilayer. Proc.Natl.Acad.Sci.Usa V. 111 3865 2014.
ISSN: ISSN 0027-8424
PubMed: 24613931
DOI: 10.1073/PNAS.1324245111
Page generated: Tue Aug 5 12:35:28 2025
ISSN: ISSN 0027-8424
PubMed: 24613931
DOI: 10.1073/PNAS.1324245111
Last articles
Fe in 5XVCFe in 5XY4
Fe in 5XX9
Fe in 5XXI
Fe in 5XVZ
Fe in 5XW2
Fe in 5XVB
Fe in 5XMJ
Fe in 5XTD
Fe in 5XTB