Iron »
PDB 4q4u-4r21 »
4qi3 »
Iron in PDB 4qi3: Cytochrome Domain of Myriococcum Thermophilum Cellobiose Dehydrogenase, Mtcyt
Protein crystallography data
The structure of Cytochrome Domain of Myriococcum Thermophilum Cellobiose Dehydrogenase, Mtcyt, PDB code: 4qi3
was solved by
T.C.Tan,
R.Gandini,
C.Sygmund,
R.Kittl,
D.Haltrich,
R.Ludwig,
B.M.Hallberg,
C.Divne,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 44.06 / 1.40 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 49.405, 56.357, 73.005, 90.00, 104.56, 90.00 |
| R / Rfree (%) | 17.8 / 22.8 |
Other elements in 4qi3:
The structure of Cytochrome Domain of Myriococcum Thermophilum Cellobiose Dehydrogenase, Mtcyt also contains other interesting chemical elements:
| Magnesium | (Mg) | 3 atoms |
Iron Binding Sites:
The binding sites of Iron atom in the Cytochrome Domain of Myriococcum Thermophilum Cellobiose Dehydrogenase, Mtcyt
(pdb code 4qi3). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 2 binding sites of Iron where determined in the Cytochrome Domain of Myriococcum Thermophilum Cellobiose Dehydrogenase, Mtcyt, PDB code: 4qi3:
Jump to Iron binding site number: 1; 2;
In total 2 binding sites of Iron where determined in the Cytochrome Domain of Myriococcum Thermophilum Cellobiose Dehydrogenase, Mtcyt, PDB code: 4qi3:
Jump to Iron binding site number: 1; 2;
Iron binding site 1 out of 2 in 4qi3
Go back to
Iron binding site 1 out
of 2 in the Cytochrome Domain of Myriococcum Thermophilum Cellobiose Dehydrogenase, Mtcyt
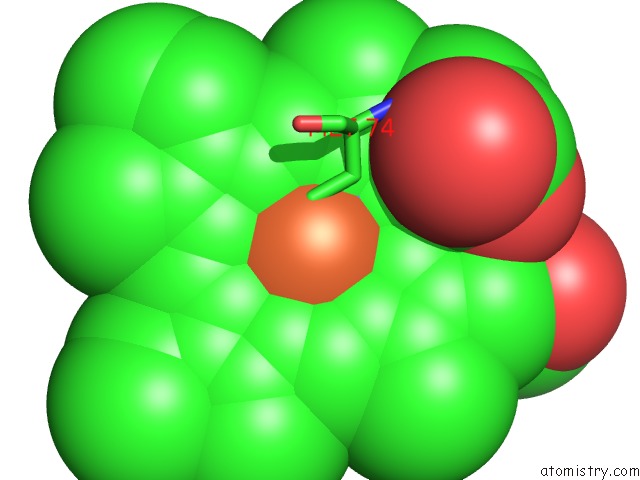
Mono view
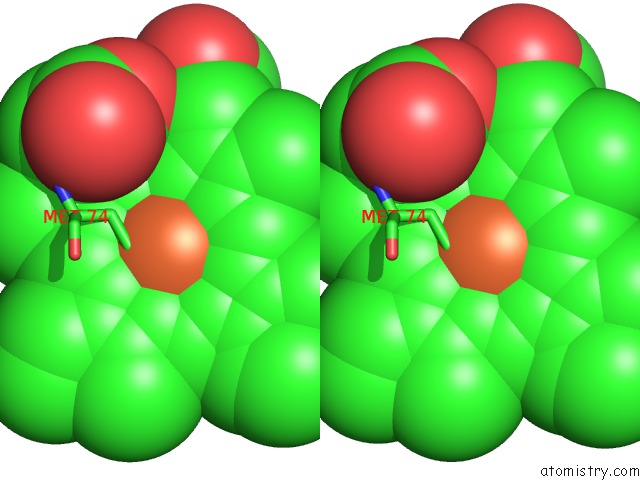
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Cytochrome Domain of Myriococcum Thermophilum Cellobiose Dehydrogenase, Mtcyt within 5.0Å range:
|
Iron binding site 2 out of 2 in 4qi3
Go back to
Iron binding site 2 out
of 2 in the Cytochrome Domain of Myriococcum Thermophilum Cellobiose Dehydrogenase, Mtcyt
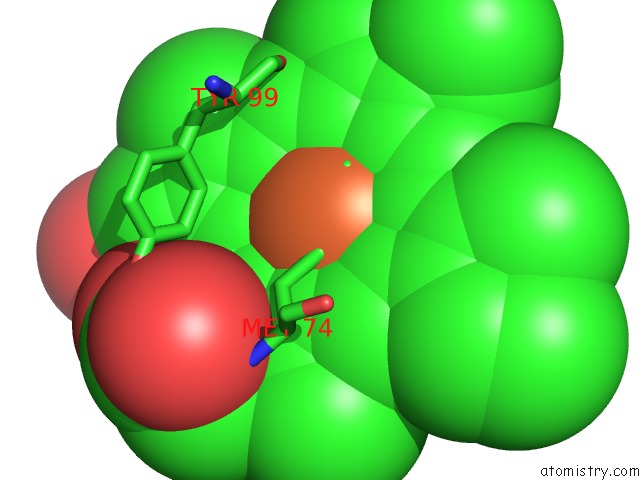
Mono view
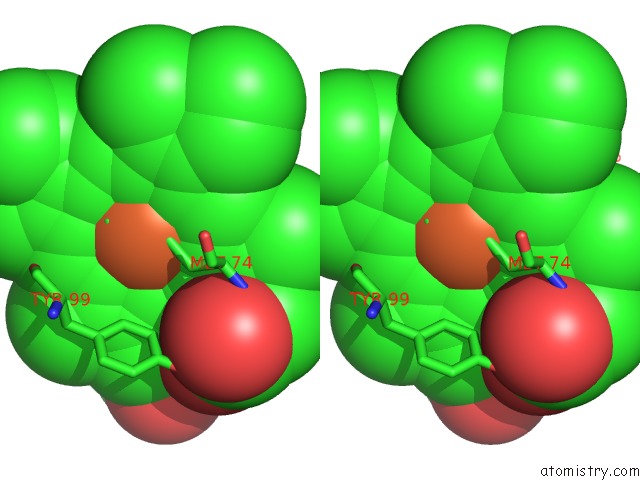
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Cytochrome Domain of Myriococcum Thermophilum Cellobiose Dehydrogenase, Mtcyt within 5.0Å range:
|
Reference:
T.C.Tan,
D.Kracher,
R.Gandini,
C.Sygmund,
R.Kittl,
D.Haltrich,
B.M.Hallberg,
R.Ludwig,
C.Divne.
Structural Basis For Cellobiose Dehydrogenase Action During Oxidative Cellulose Degradation. Nat Commun V. 6 7542 2015.
ISSN: ESSN 2041-1723
PubMed: 26151670
DOI: 10.1038/NCOMMS8542
Page generated: Tue Aug 5 13:50:52 2025
ISSN: ESSN 2041-1723
PubMed: 26151670
DOI: 10.1038/NCOMMS8542
Last articles
Fe in 4TODFe in 4TO9
Fe in 4TOB
Fe in 4TKV
Fe in 4TKU
Fe in 4TNK
Fe in 4TNJ
Fe in 4TNI
Fe in 4TNH
Fe in 4TLF