Iron »
PDB 5kts-5lg8 »
5l8d »
Iron in PDB 5l8d: X-Ray Structure of Nika From Escherichia Coli in Complex with Ru(Bis(Pyrzol-1-Yl)Acetate Scorpionate)(Co)2CL
Protein crystallography data
The structure of X-Ray Structure of Nika From Escherichia Coli in Complex with Ru(Bis(Pyrzol-1-Yl)Acetate Scorpionate)(Co)2CL, PDB code: 5l8d
was solved by
S.Menage,
C.Cavazza,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 46.80 / 1.80 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 86.303, 93.606, 124.211, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 16.8 / 20.8 |
Other elements in 5l8d:
The structure of X-Ray Structure of Nika From Escherichia Coli in Complex with Ru(Bis(Pyrzol-1-Yl)Acetate Scorpionate)(Co)2CL also contains other interesting chemical elements:
| Ruthenium | (Ru) | 1 atom |
| Chlorine | (Cl) | 2 atoms |
Iron Binding Sites:
The binding sites of Iron atom in the X-Ray Structure of Nika From Escherichia Coli in Complex with Ru(Bis(Pyrzol-1-Yl)Acetate Scorpionate)(Co)2CL
(pdb code 5l8d). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total only one binding site of Iron was determined in the X-Ray Structure of Nika From Escherichia Coli in Complex with Ru(Bis(Pyrzol-1-Yl)Acetate Scorpionate)(Co)2CL, PDB code: 5l8d:
In total only one binding site of Iron was determined in the X-Ray Structure of Nika From Escherichia Coli in Complex with Ru(Bis(Pyrzol-1-Yl)Acetate Scorpionate)(Co)2CL, PDB code: 5l8d:
Iron binding site 1 out of 1 in 5l8d
Go back to
Iron binding site 1 out
of 1 in the X-Ray Structure of Nika From Escherichia Coli in Complex with Ru(Bis(Pyrzol-1-Yl)Acetate Scorpionate)(Co)2CL
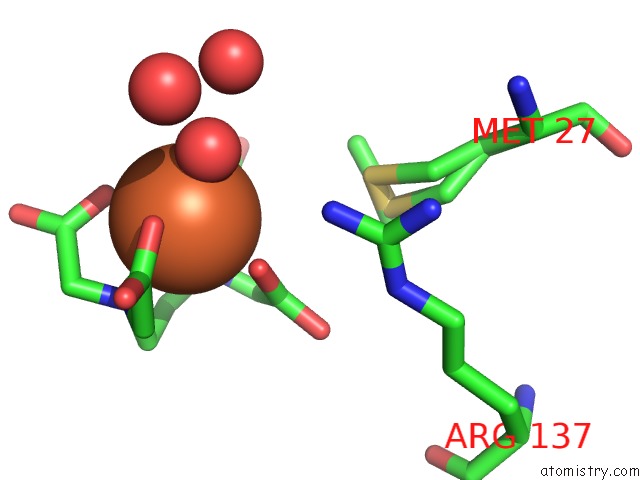
Mono view
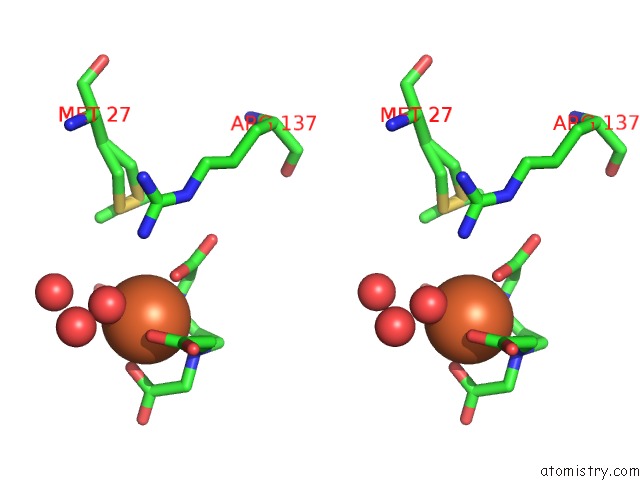
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of X-Ray Structure of Nika From Escherichia Coli in Complex with Ru(Bis(Pyrzol-1-Yl)Acetate Scorpionate)(Co)2CL within 5.0Å range:
|
Reference:
S.Lopez,
L.Rondot,
C.Cavazza,
M.Iannello,
E.Boeri-Erba,
N.Burzlaff,
F.Strinitz,
A.Jorge-Robin,
C.Marchi-Delapierre,
S.Menage.
Efficient Conversion of Alkenes to Chlorohydrins By A Ru-Based Artificial Enzyme. Chem. Commun. (Camb.) V. 53 3579 2017.
ISSN: ESSN 1364-548X
PubMed: 28289745
DOI: 10.1039/C6CC08873B
Page generated: Tue Aug 6 04:06:23 2024
ISSN: ESSN 1364-548X
PubMed: 28289745
DOI: 10.1039/C6CC08873B
Last articles
F in 7NVMF in 7NW2
F in 7NW0
F in 7NVL
F in 7NVX
F in 7NVV
F in 7NVO
F in 7NTH
F in 7NTI
F in 7NPC