Iron »
PDB 5sx3-5tia »
5t5v »
Iron in PDB 5t5v: Lipoxygenase-1 (Soybean) at 293K
Enzymatic activity of Lipoxygenase-1 (Soybean) at 293K
All present enzymatic activity of Lipoxygenase-1 (Soybean) at 293K:
1.13.11.12;
1.13.11.12;
Protein crystallography data
The structure of Lipoxygenase-1 (Soybean) at 293K, PDB code: 5t5v
was solved by
E.M.Poss,
J.S.Fraser,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 91.38 / 1.80 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 91.572, 92.754, 100.919, 90.00, 93.76, 90.00 |
| R / Rfree (%) | 13.4 / 17.1 |
Iron Binding Sites:
The binding sites of Iron atom in the Lipoxygenase-1 (Soybean) at 293K
(pdb code 5t5v). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 2 binding sites of Iron where determined in the Lipoxygenase-1 (Soybean) at 293K, PDB code: 5t5v:
Jump to Iron binding site number: 1; 2;
In total 2 binding sites of Iron where determined in the Lipoxygenase-1 (Soybean) at 293K, PDB code: 5t5v:
Jump to Iron binding site number: 1; 2;
Iron binding site 1 out of 2 in 5t5v
Go back to
Iron binding site 1 out
of 2 in the Lipoxygenase-1 (Soybean) at 293K
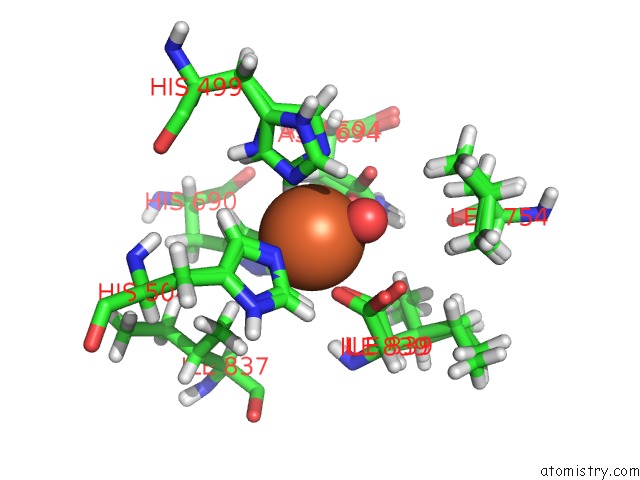
Mono view
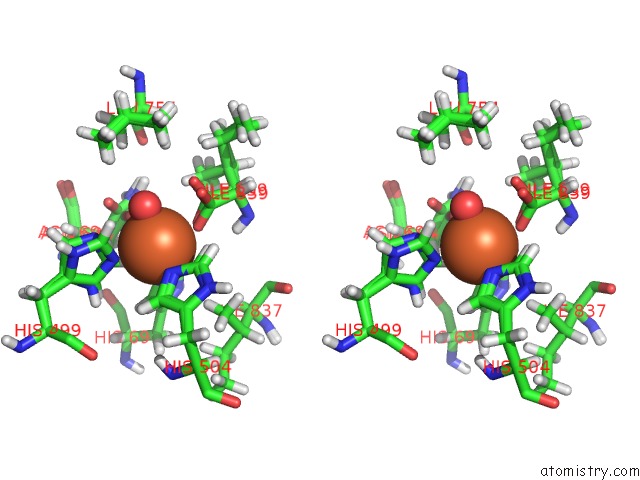
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Lipoxygenase-1 (Soybean) at 293K within 5.0Å range:
|
Iron binding site 2 out of 2 in 5t5v
Go back to
Iron binding site 2 out
of 2 in the Lipoxygenase-1 (Soybean) at 293K
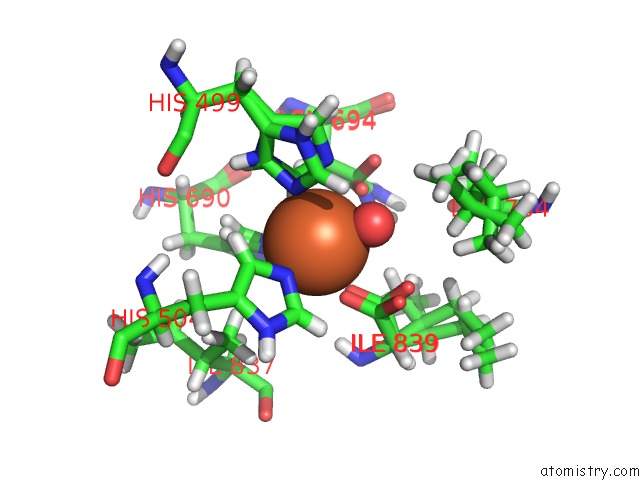
Mono view
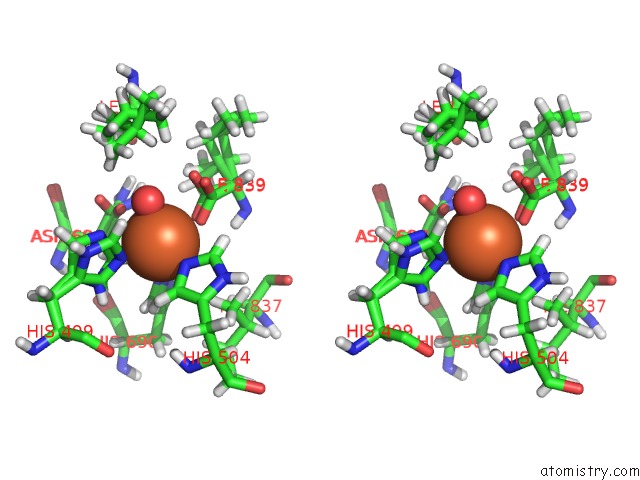
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Lipoxygenase-1 (Soybean) at 293K within 5.0Å range:
|
Reference:
A.R.Offenbacher,
S.Hu,
E.M.Poss,
C.A.M.Carr,
A.D.Scouras,
D.M.Prigozhin,
A.T.Iavarone,
A.Palla,
T.Alber,
J.S.Fraser,
J.P.Klinman.
Hydrogen-Deuterium Exchange of Lipoxygenase Uncovers A Relationship Between Distal, Solvent Exposed Protein Motions and the Thermal Activation Barrier For Catalytic Proton-Coupled Electron Tunneling. Acs Cent Sci V. 3 570 2017.
ISSN: ESSN 2374-7943
PubMed: 28691068
DOI: 10.1021/ACSCENTSCI.7B00142
Page generated: Tue Aug 6 08:53:29 2024
ISSN: ESSN 2374-7943
PubMed: 28691068
DOI: 10.1021/ACSCENTSCI.7B00142
Last articles
F in 4LXKF in 4LXI
F in 4LX0
F in 4LXB
F in 4LVT
F in 4LWV
F in 4LW1
F in 4LWU
F in 4LTS
F in 4LUD