Iron »
PDB 5xzi-5ylw »
5ybr »
Iron in PDB 5ybr: Fe(II)/(Alpha)Ketoglutarate-Dependent Dioxygenase Prha-V150L/A232S in Complex with Preaustinoid A3
Protein crystallography data
The structure of Fe(II)/(Alpha)Ketoglutarate-Dependent Dioxygenase Prha-V150L/A232S in Complex with Preaustinoid A3, PDB code: 5ybr
was solved by
Y.Nakashima,
M.Senda,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 43.67 / 2.26 |
| Space group | P 62 |
| Cell size a, b, c (Å), α, β, γ (°) | 171.382, 171.382, 45.696, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 21.2 / 25.9 |
Iron Binding Sites:
The binding sites of Iron atom in the Fe(II)/(Alpha)Ketoglutarate-Dependent Dioxygenase Prha-V150L/A232S in Complex with Preaustinoid A3
(pdb code 5ybr). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 2 binding sites of Iron where determined in the Fe(II)/(Alpha)Ketoglutarate-Dependent Dioxygenase Prha-V150L/A232S in Complex with Preaustinoid A3, PDB code: 5ybr:
Jump to Iron binding site number: 1; 2;
In total 2 binding sites of Iron where determined in the Fe(II)/(Alpha)Ketoglutarate-Dependent Dioxygenase Prha-V150L/A232S in Complex with Preaustinoid A3, PDB code: 5ybr:
Jump to Iron binding site number: 1; 2;
Iron binding site 1 out of 2 in 5ybr
Go back to
Iron binding site 1 out
of 2 in the Fe(II)/(Alpha)Ketoglutarate-Dependent Dioxygenase Prha-V150L/A232S in Complex with Preaustinoid A3

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Fe(II)/(Alpha)Ketoglutarate-Dependent Dioxygenase Prha-V150L/A232S in Complex with Preaustinoid A3 within 5.0Å range:
|
Iron binding site 2 out of 2 in 5ybr
Go back to
Iron binding site 2 out
of 2 in the Fe(II)/(Alpha)Ketoglutarate-Dependent Dioxygenase Prha-V150L/A232S in Complex with Preaustinoid A3

Mono view
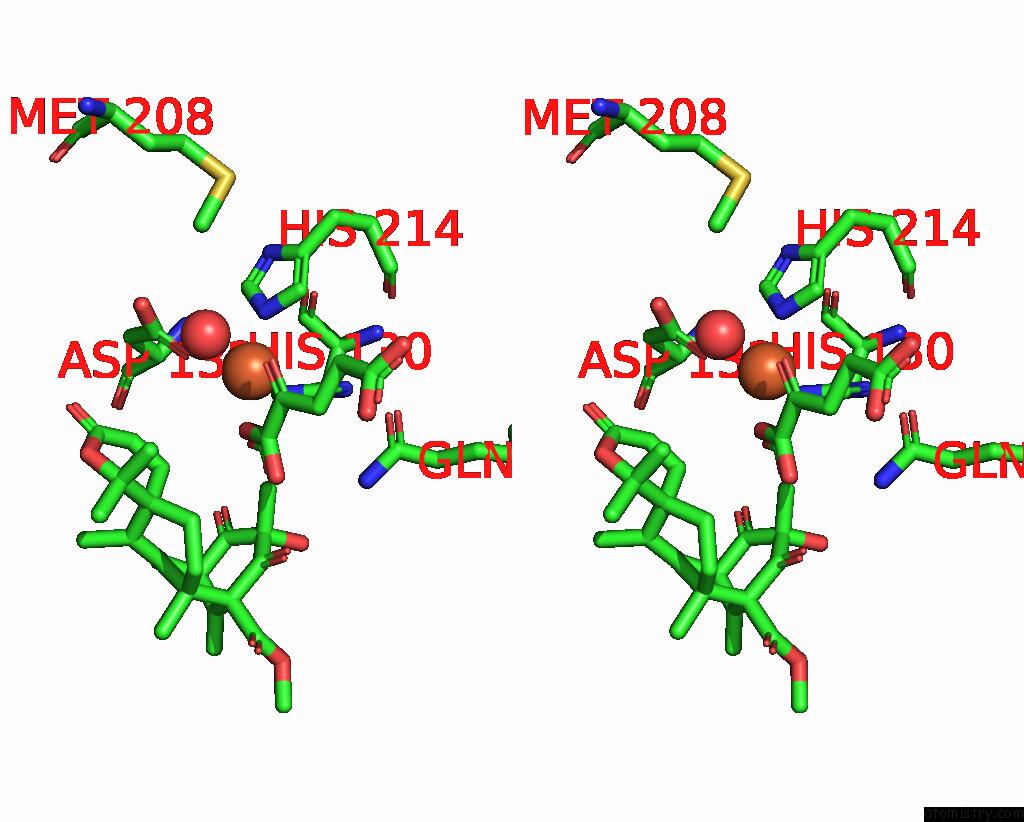
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Fe(II)/(Alpha)Ketoglutarate-Dependent Dioxygenase Prha-V150L/A232S in Complex with Preaustinoid A3 within 5.0Å range:
|
Reference:
Y.Nakashima,
T.Mori,
H.Nakamura,
T.Awakawa,
S.Hoshino,
M.Senda,
T.Senda,
I.Abe.
Structure Function and Engineering of Multifunctional Non-Heme Iron Dependent Oxygenases in Fungal Meroterpenoid Biosynthesis. Nat Commun V. 9 104 2018.
ISSN: ESSN 2041-1723
PubMed: 29317628
DOI: 10.1038/S41467-017-02371-W
Page generated: Tue Aug 6 12:46:58 2024
ISSN: ESSN 2041-1723
PubMed: 29317628
DOI: 10.1038/S41467-017-02371-W
Last articles
F in 4Q09F in 4PKO
F in 4PKN
F in 4Q06
F in 4Q02
F in 4PYY
F in 4PZH
F in 4PYX
F in 4PX5
F in 4PY4