Iron »
PDB 6e45-6f0a »
6e4v »
Iron in PDB 6e4v: The Crystal Structure of Fhue From E. Coli in Complex with Its Substrate Coprogen
Protein crystallography data
The structure of The Crystal Structure of Fhue From E. Coli in Complex with Its Substrate Coprogen, PDB code: 6e4v
was solved by
R.Grinter,
T.Lithgow,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 47.57 / 2.00 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 68.190, 103.860, 118.620, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.6 / 23.5 |
Iron Binding Sites:
The binding sites of Iron atom in the The Crystal Structure of Fhue From E. Coli in Complex with Its Substrate Coprogen
(pdb code 6e4v). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total only one binding site of Iron was determined in the The Crystal Structure of Fhue From E. Coli in Complex with Its Substrate Coprogen, PDB code: 6e4v:
In total only one binding site of Iron was determined in the The Crystal Structure of Fhue From E. Coli in Complex with Its Substrate Coprogen, PDB code: 6e4v:
Iron binding site 1 out of 1 in 6e4v
Go back to
Iron binding site 1 out
of 1 in the The Crystal Structure of Fhue From E. Coli in Complex with Its Substrate Coprogen
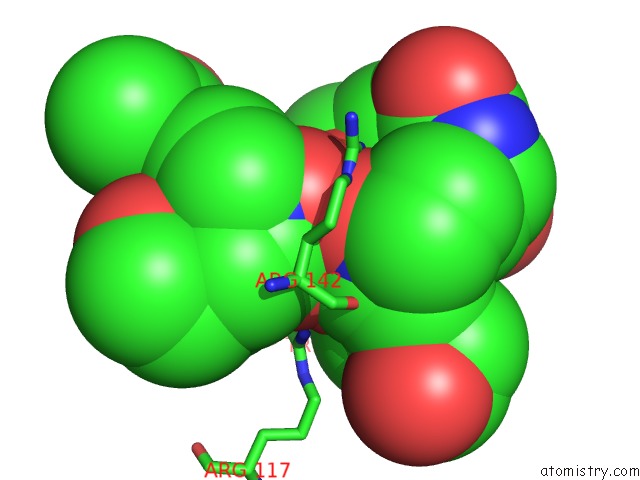
Mono view
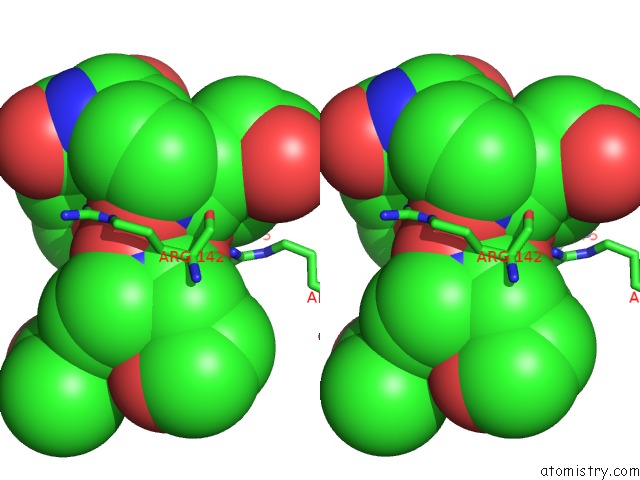
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of The Crystal Structure of Fhue From E. Coli in Complex with Its Substrate Coprogen within 5.0Å range:
|
Reference:
R.Grinter,
T.Lithgow.
Determination of the Molecular Basis For Coprogen Import By Gram-Negative Bacteria. Iucrj V. 6 401 2019.
ISSN: ESSN 2052-2525
PubMed: 31098021
DOI: 10.1107/S2052252519002926
Page generated: Tue Aug 6 17:18:14 2024
ISSN: ESSN 2052-2525
PubMed: 31098021
DOI: 10.1107/S2052252519002926
Last articles
Cl in 2YHICl in 2YIH
Cl in 2YI7
Cl in 2YI5
Cl in 2YI0
Cl in 2YGY
Cl in 2YHY
Cl in 2YHW
Cl in 2YG9
Cl in 2YHG