Iron »
PDB 7k5g-7kvo »
7kc0 »
Iron in PDB 7kc0: Structure of the Saccharomyces Cerevisiae Replicative Polymerase Delta in Complex with A Primer/Template and the Pcna Clamp
Enzymatic activity of Structure of the Saccharomyces Cerevisiae Replicative Polymerase Delta in Complex with A Primer/Template and the Pcna Clamp
All present enzymatic activity of Structure of the Saccharomyces Cerevisiae Replicative Polymerase Delta in Complex with A Primer/Template and the Pcna Clamp:
2.7.7.7;
2.7.7.7;
Other elements in 7kc0:
The structure of Structure of the Saccharomyces Cerevisiae Replicative Polymerase Delta in Complex with A Primer/Template and the Pcna Clamp also contains other interesting chemical elements:
| Magnesium | (Mg) | 2 atoms |
| Zinc | (Zn) | 1 atom |
Iron Binding Sites:
The binding sites of Iron atom in the Structure of the Saccharomyces Cerevisiae Replicative Polymerase Delta in Complex with A Primer/Template and the Pcna Clamp
(pdb code 7kc0). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 4 binding sites of Iron where determined in the Structure of the Saccharomyces Cerevisiae Replicative Polymerase Delta in Complex with A Primer/Template and the Pcna Clamp, PDB code: 7kc0:
Jump to Iron binding site number: 1; 2; 3; 4;
In total 4 binding sites of Iron where determined in the Structure of the Saccharomyces Cerevisiae Replicative Polymerase Delta in Complex with A Primer/Template and the Pcna Clamp, PDB code: 7kc0:
Jump to Iron binding site number: 1; 2; 3; 4;
Iron binding site 1 out of 4 in 7kc0
Go back to
Iron binding site 1 out
of 4 in the Structure of the Saccharomyces Cerevisiae Replicative Polymerase Delta in Complex with A Primer/Template and the Pcna Clamp

Mono view
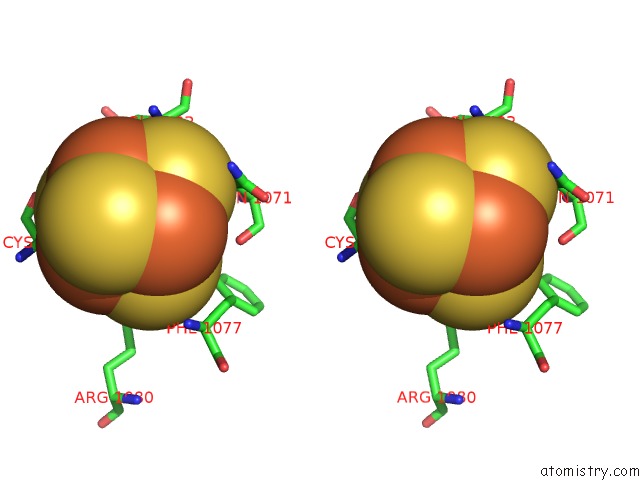
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Structure of the Saccharomyces Cerevisiae Replicative Polymerase Delta in Complex with A Primer/Template and the Pcna Clamp within 5.0Å range:
|
Iron binding site 2 out of 4 in 7kc0
Go back to
Iron binding site 2 out
of 4 in the Structure of the Saccharomyces Cerevisiae Replicative Polymerase Delta in Complex with A Primer/Template and the Pcna Clamp
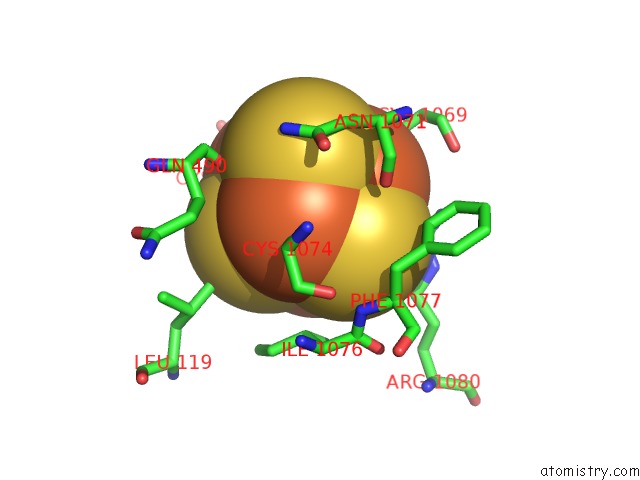
Mono view
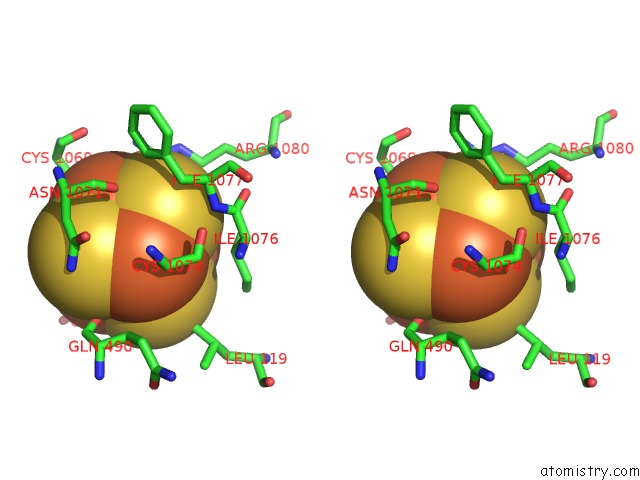
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Structure of the Saccharomyces Cerevisiae Replicative Polymerase Delta in Complex with A Primer/Template and the Pcna Clamp within 5.0Å range:
|
Iron binding site 3 out of 4 in 7kc0
Go back to
Iron binding site 3 out
of 4 in the Structure of the Saccharomyces Cerevisiae Replicative Polymerase Delta in Complex with A Primer/Template and the Pcna Clamp

Mono view
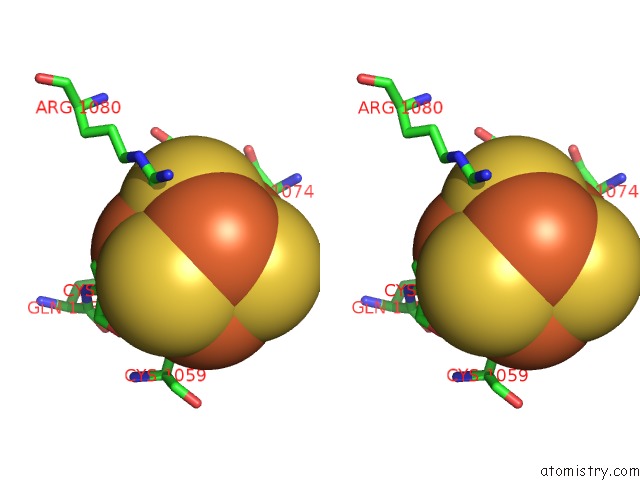
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 3 of Structure of the Saccharomyces Cerevisiae Replicative Polymerase Delta in Complex with A Primer/Template and the Pcna Clamp within 5.0Å range:
|
Iron binding site 4 out of 4 in 7kc0
Go back to
Iron binding site 4 out
of 4 in the Structure of the Saccharomyces Cerevisiae Replicative Polymerase Delta in Complex with A Primer/Template and the Pcna Clamp
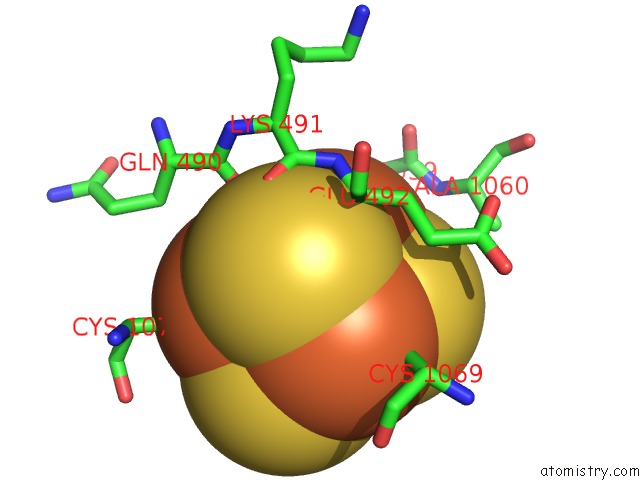
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 4 of Structure of the Saccharomyces Cerevisiae Replicative Polymerase Delta in Complex with A Primer/Template and the Pcna Clamp within 5.0Å range:
|
Reference:
F.Zheng,
R.E.Georgescu,
H.Li,
M.E.O'donnell.
Structure of Eukaryotic Dna Polymerase Delta Bound to the Pcna Clamp While Encircling Dna. Proc.Natl.Acad.Sci.Usa 2020.
ISSN: ESSN 1091-6490
PubMed: 33203675
DOI: 10.1073/PNAS.2017637117
Page generated: Thu Aug 8 06:23:58 2024
ISSN: ESSN 1091-6490
PubMed: 33203675
DOI: 10.1073/PNAS.2017637117
Last articles
F in 7O6IF in 7O4J
F in 7O4I
F in 7O4L
F in 7O4K
F in 7O2V
F in 7NZX
F in 7O2R
F in 7NZW
F in 7O2F