Iron »
PDB 7ni3-7o4i »
7npa »
Iron in PDB 7npa: Crystal Structure of the Coenzyme F420-Dependent Sulfite Reductase From Methanothermococcus Thermolithotrophicus at 1.55-A Resolution
Enzymatic activity of Crystal Structure of the Coenzyme F420-Dependent Sulfite Reductase From Methanothermococcus Thermolithotrophicus at 1.55-A Resolution
All present enzymatic activity of Crystal Structure of the Coenzyme F420-Dependent Sulfite Reductase From Methanothermococcus Thermolithotrophicus at 1.55-A Resolution:
1.8.98.3;
1.8.98.3;
Protein crystallography data
The structure of Crystal Structure of the Coenzyme F420-Dependent Sulfite Reductase From Methanothermococcus Thermolithotrophicus at 1.55-A Resolution, PDB code: 7npa
was solved by
M.Jespersen,
T.Wagner,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 77.29 / 1.55 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 113.148, 124.157, 241.062, 102.28, 95.71, 90.25 |
| R / Rfree (%) | 15.9 / 17.1 |
Other elements in 7npa:
The structure of Crystal Structure of the Coenzyme F420-Dependent Sulfite Reductase From Methanothermococcus Thermolithotrophicus at 1.55-A Resolution also contains other interesting chemical elements:
| Chlorine | (Cl) | 29 atoms |
Iron Binding Sites:
Pages:
>>> Page 1 <<< Page 2, Binding sites: 11 - 20; Page 3, Binding sites: 21 - 30; Page 4, Binding sites: 31 - 40; Page 5, Binding sites: 41 - 50; Page 6, Binding sites: 51 - 60; Page 7, Binding sites: 61 - 70; Page 8, Binding sites: 71 - 80; Page 9, Binding sites: 81 - 90; Page 10, Binding sites: 91 - 100; Page 11, Binding sites: 101 - 110; Page 12, Binding sites: 111 - 120; Page 13, Binding sites: 121 - 130; Page 14, Binding sites: 131 - 140; Page 15, Binding sites: 141 - 150; Page 16, Binding sites: 151 - 160; Page 17, Binding sites: 161 - 170; Page 18, Binding sites: 171 - 180; Page 19, Binding sites: 181 - 190; Page 20, Binding sites: 191 - 200; Page 21, Binding sites: 201 - 210; Page 22, Binding sites: 211 - 220; Page 23, Binding sites: 221 - 230; Page 24, Binding sites: 231 - 240; Page 25, Binding sites: 241 - 250; Page 26, Binding sites: 251 - 260; Page 27, Binding sites: 261 - 270; Page 28, Binding sites: 271 - 280; Page 29, Binding sites: 281 - 290; Page 30, Binding sites: 291 - 300; Page 31, Binding sites: 301 - 310; Page 32, Binding sites: 311 - 320; Page 33, Binding sites: 321 - 330; Page 34, Binding sites: 331 - 340; Page 35, Binding sites: 341 - 350; Page 36, Binding sites: 351 - 360; Page 37, Binding sites: 361 - 370; Page 38, Binding sites: 371 - 380; Page 39, Binding sites: 381 - 390; Page 40, Binding sites: 391 - 400; Page 41, Binding sites: 401 - 408;Binding sites:
The binding sites of Iron atom in the Crystal Structure of the Coenzyme F420-Dependent Sulfite Reductase From Methanothermococcus Thermolithotrophicus at 1.55-A Resolution (pdb code 7npa). This binding sites where shown within 5.0 Angstroms radius around Iron atom.In total 408 binding sites of Iron where determined in the Crystal Structure of the Coenzyme F420-Dependent Sulfite Reductase From Methanothermococcus Thermolithotrophicus at 1.55-A Resolution, PDB code: 7npa:
Jump to Iron binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
Iron binding site 1 out of 408 in 7npa
Go back to
Iron binding site 1 out
of 408 in the Crystal Structure of the Coenzyme F420-Dependent Sulfite Reductase From Methanothermococcus Thermolithotrophicus at 1.55-A Resolution
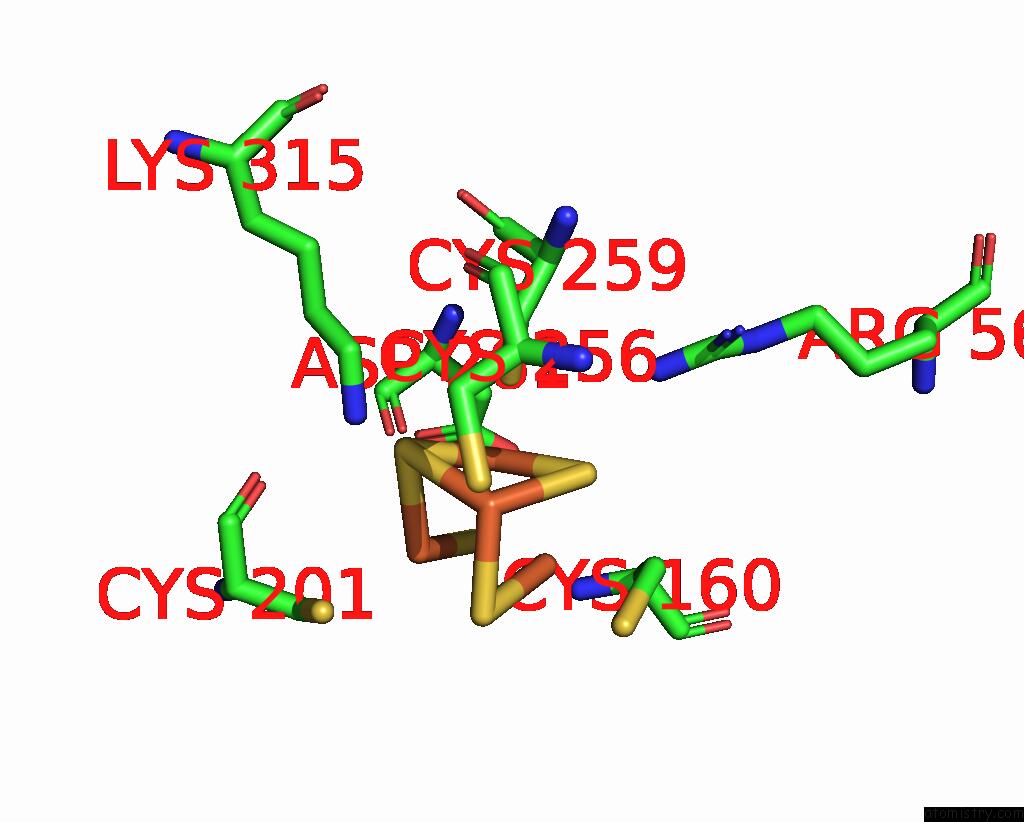
Mono view
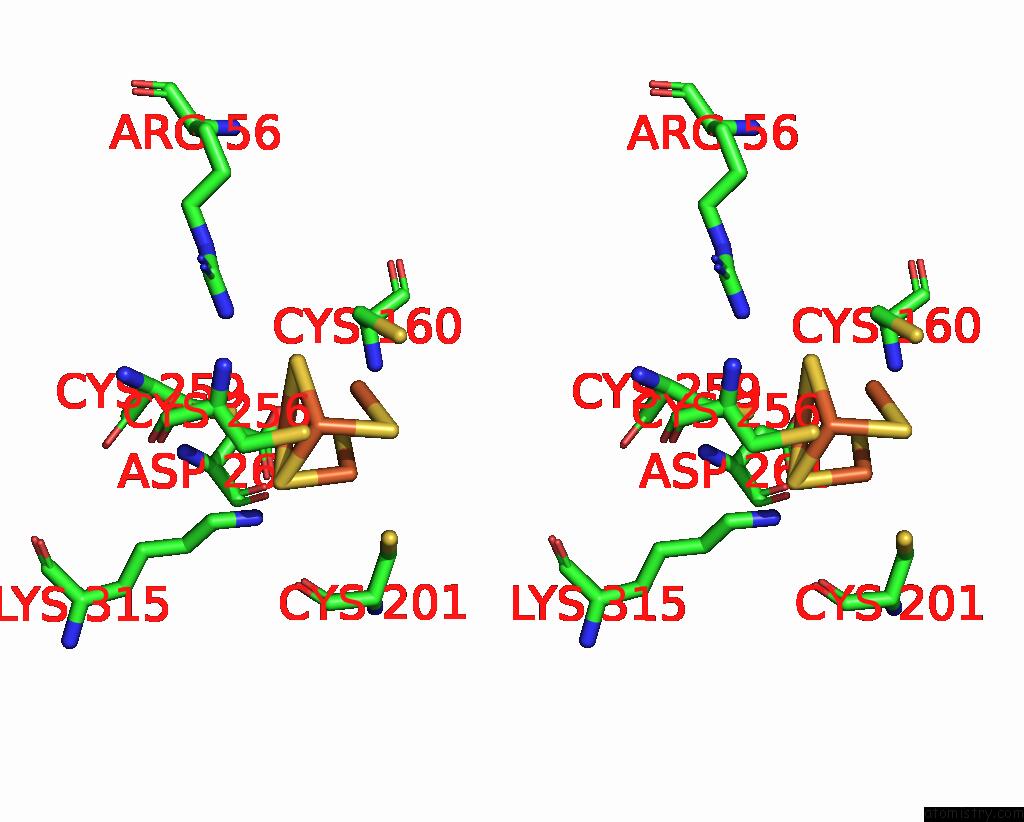
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Crystal Structure of the Coenzyme F420-Dependent Sulfite Reductase From Methanothermococcus Thermolithotrophicus at 1.55-A Resolution within 5.0Å range:
|
Iron binding site 2 out of 408 in 7npa
Go back to
Iron binding site 2 out
of 408 in the Crystal Structure of the Coenzyme F420-Dependent Sulfite Reductase From Methanothermococcus Thermolithotrophicus at 1.55-A Resolution
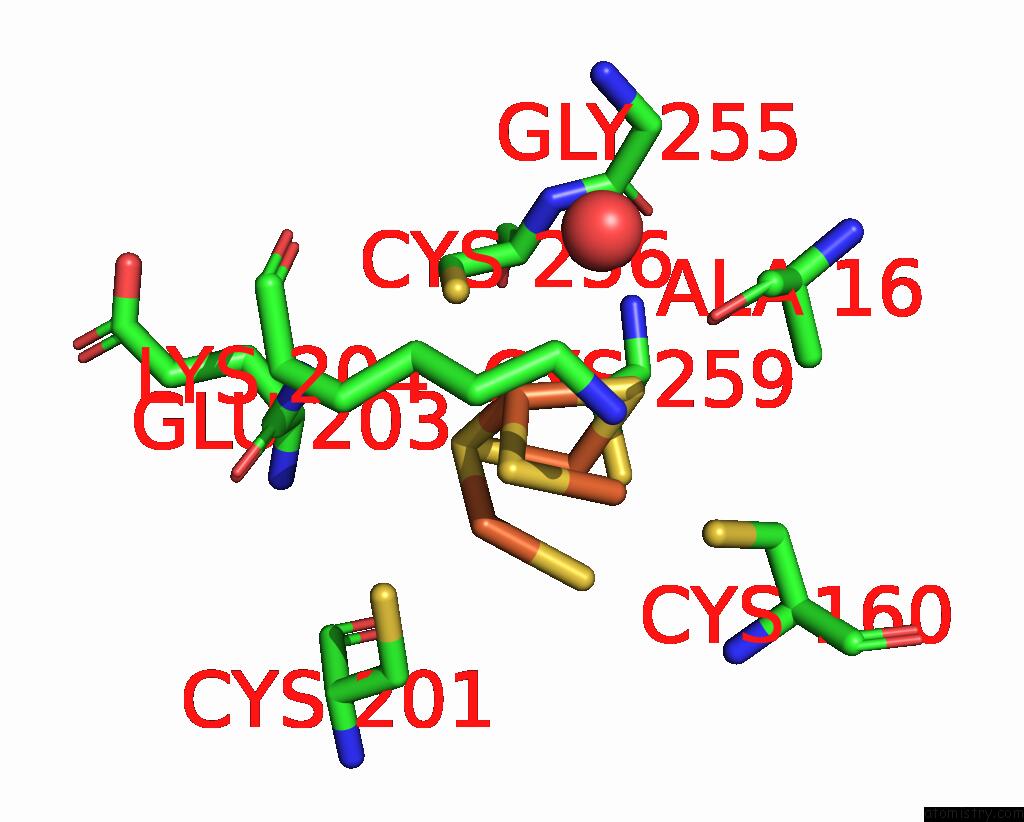
Mono view
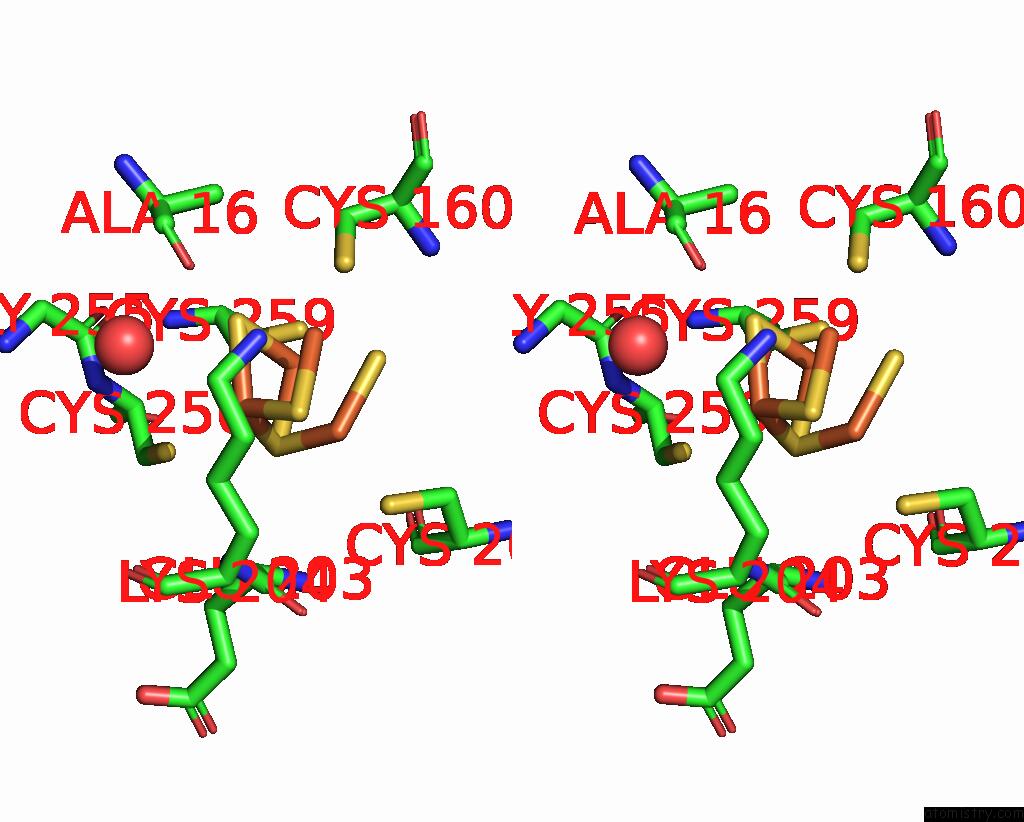
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Crystal Structure of the Coenzyme F420-Dependent Sulfite Reductase From Methanothermococcus Thermolithotrophicus at 1.55-A Resolution within 5.0Å range:
|
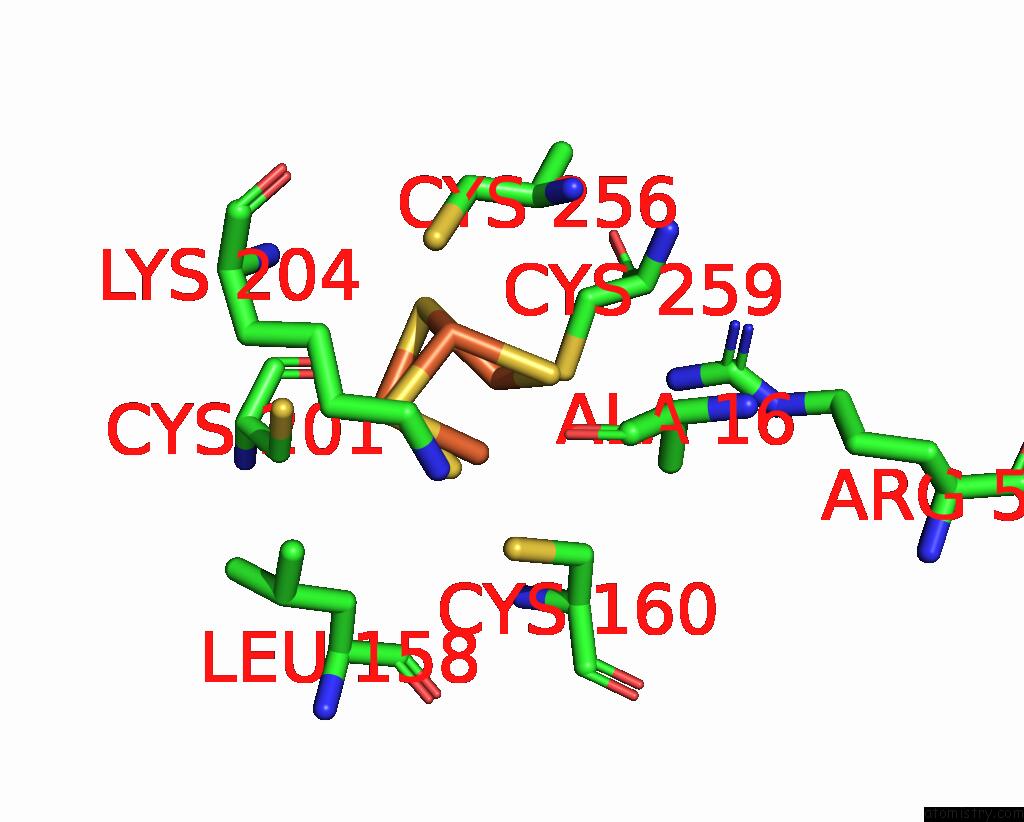
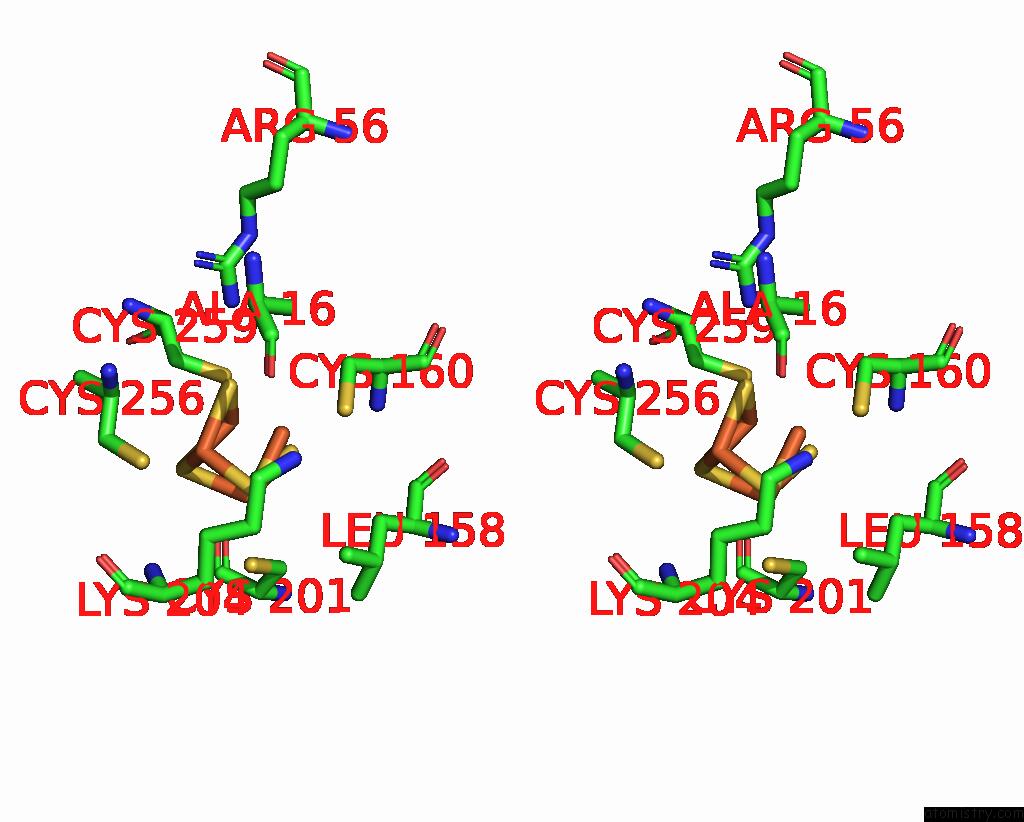
Iron binding site 3 out of 408 in 7npa
Go back to
Iron binding site 3 out
of 408 in the Crystal Structure of the Coenzyme F420-Dependent Sulfite Reductase From Methanothermococcus Thermolithotrophicus at 1.55-A Resolution
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 3 of Crystal Structure of the Coenzyme F420-Dependent Sulfite Reductase From Methanothermococcus Thermolithotrophicus at 1.55-A Resolution within 5.0Å range:
|
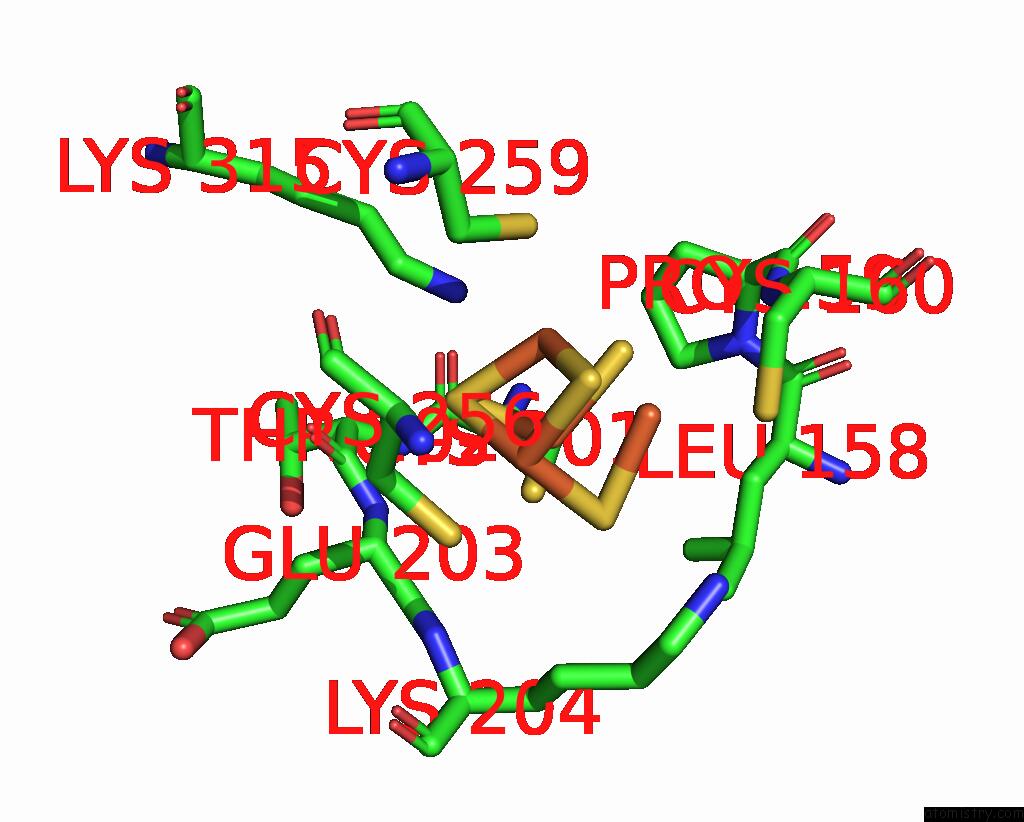
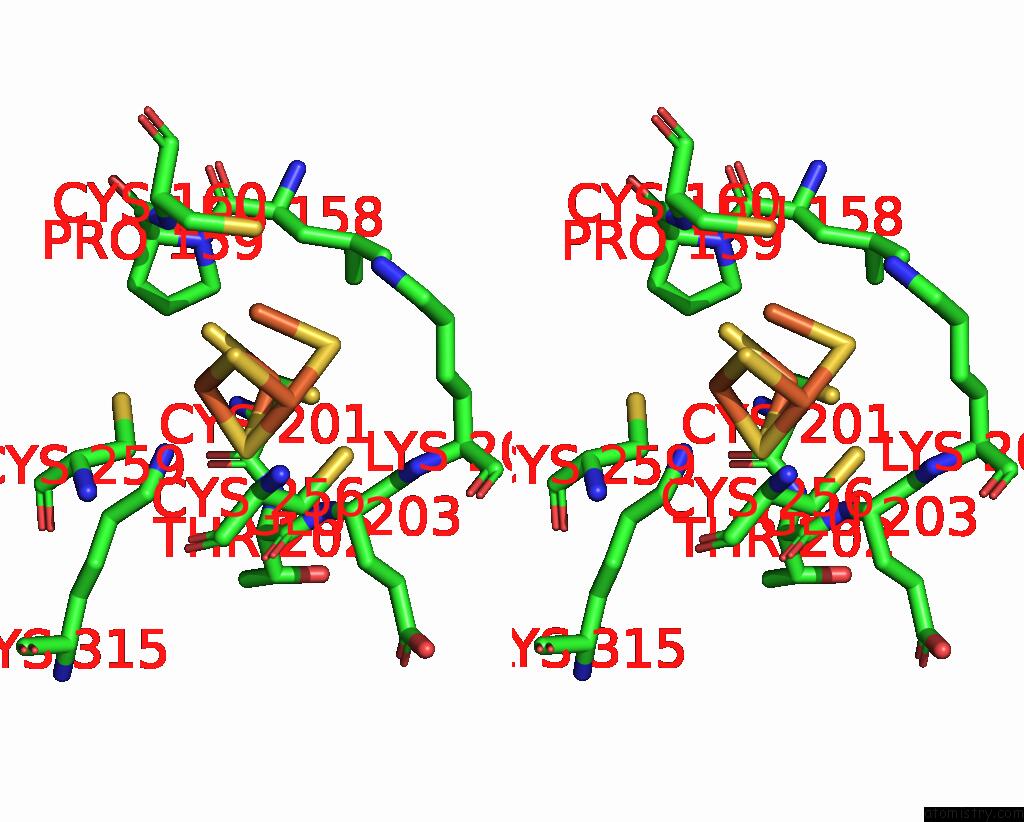
Iron binding site 4 out of 408 in 7npa
Go back to
Iron binding site 4 out
of 408 in the Crystal Structure of the Coenzyme F420-Dependent Sulfite Reductase From Methanothermococcus Thermolithotrophicus at 1.55-A Resolution
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 4 of Crystal Structure of the Coenzyme F420-Dependent Sulfite Reductase From Methanothermococcus Thermolithotrophicus at 1.55-A Resolution within 5.0Å range:
|
Iron binding site 5 out of 408 in 7npa
Go back to
Iron binding site 5 out
of 408 in the Crystal Structure of the Coenzyme F420-Dependent Sulfite Reductase From Methanothermococcus Thermolithotrophicus at 1.55-A Resolution
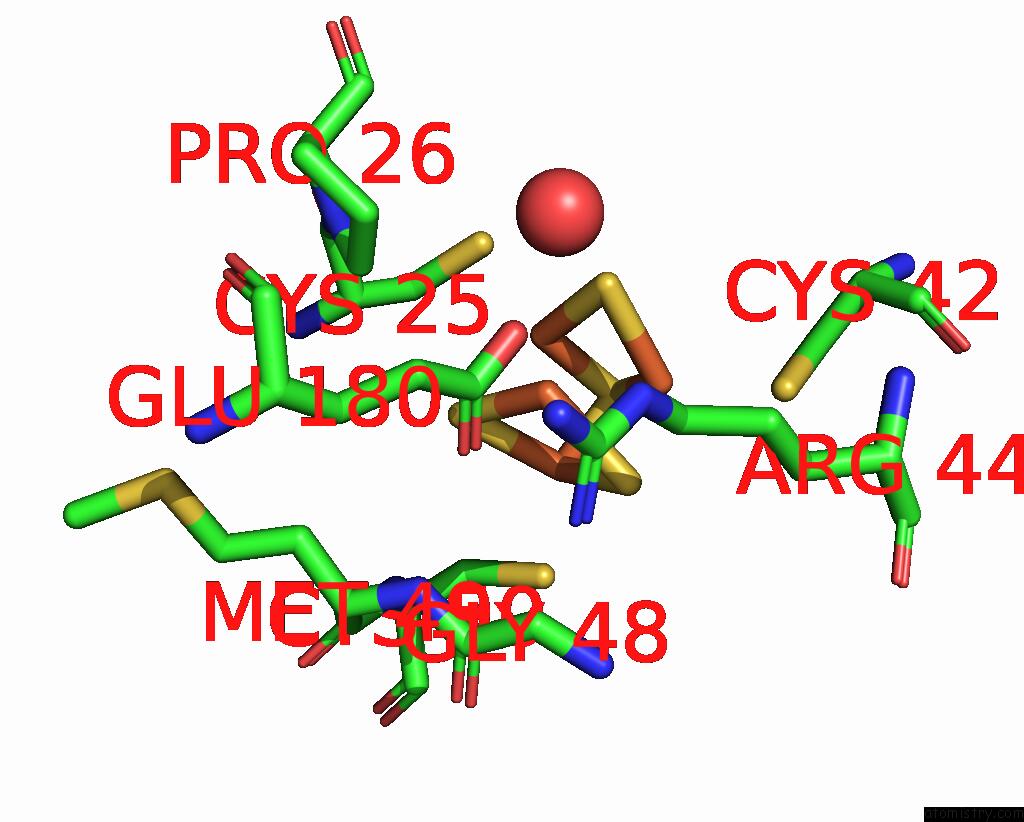
Mono view
Stereo pair view

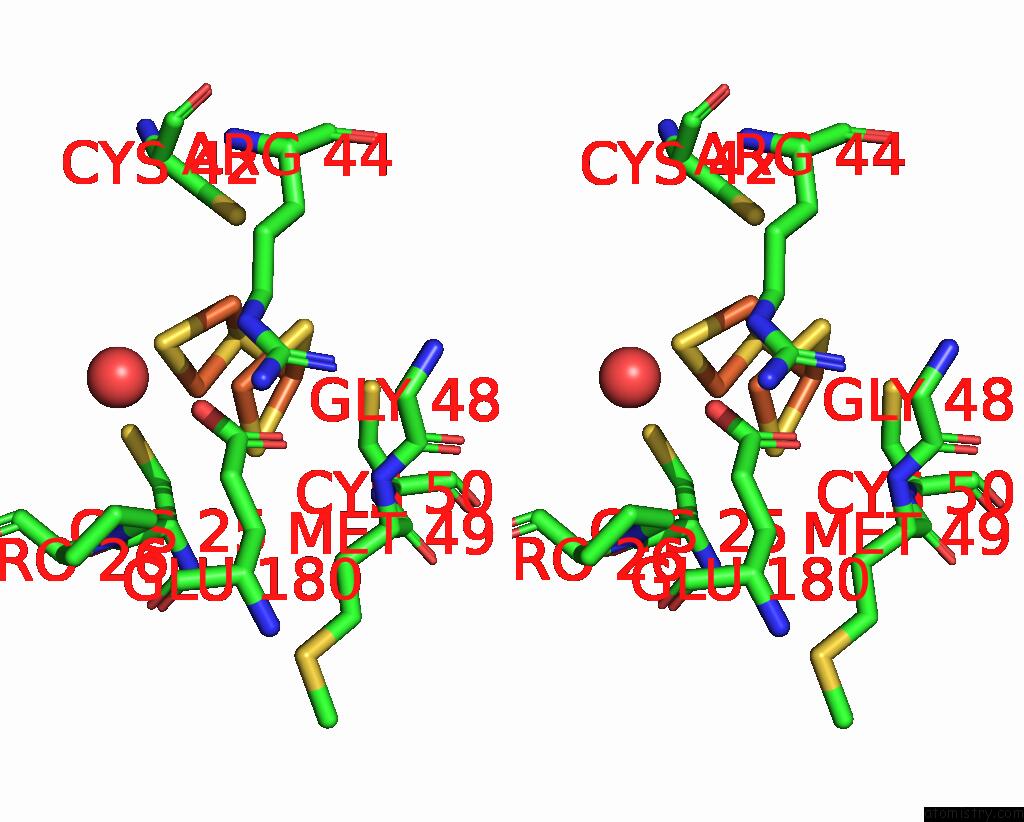
Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 5 of Crystal Structure of the Coenzyme F420-Dependent Sulfite Reductase From Methanothermococcus Thermolithotrophicus at 1.55-A Resolution within 5.0Å range:
|
Iron binding site 6 out of 408 in 7npa
Go back to
Iron binding site 6 out
of 408 in the Crystal Structure of the Coenzyme F420-Dependent Sulfite Reductase From Methanothermococcus Thermolithotrophicus at 1.55-A Resolution
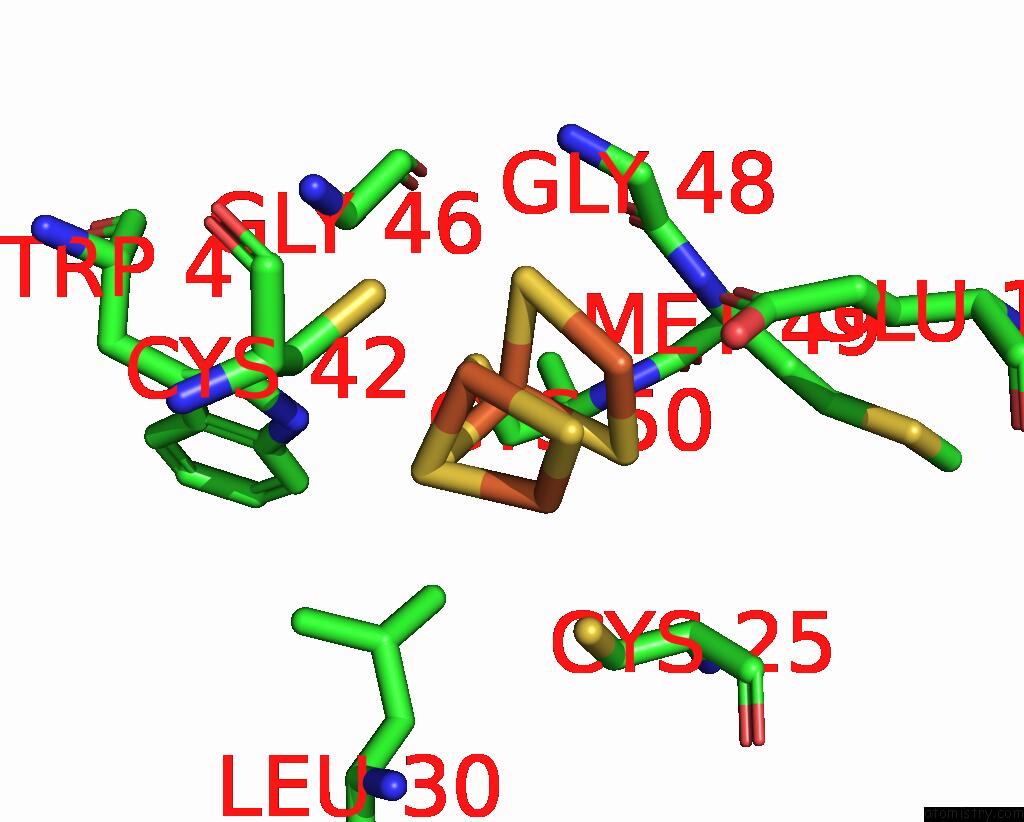
Mono view
Stereo pair view

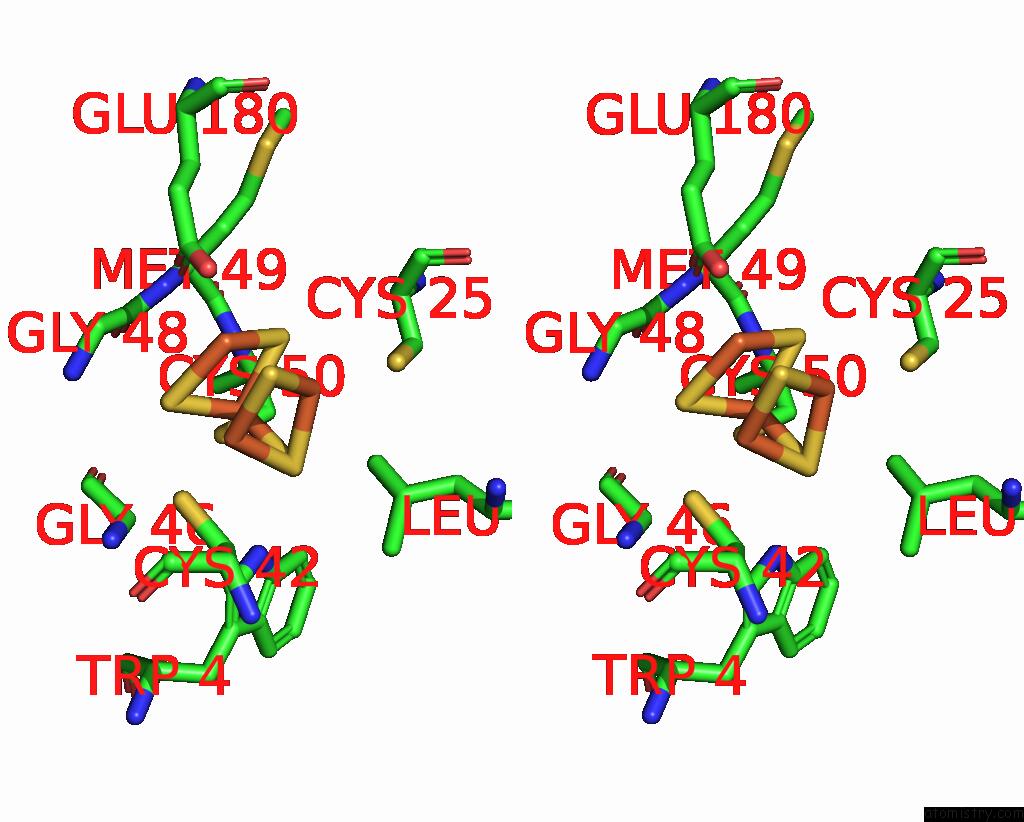
Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 6 of Crystal Structure of the Coenzyme F420-Dependent Sulfite Reductase From Methanothermococcus Thermolithotrophicus at 1.55-A Resolution within 5.0Å range:
|
Iron binding site 7 out of 408 in 7npa
Go back to
Iron binding site 7 out
of 408 in the Crystal Structure of the Coenzyme F420-Dependent Sulfite Reductase From Methanothermococcus Thermolithotrophicus at 1.55-A Resolution
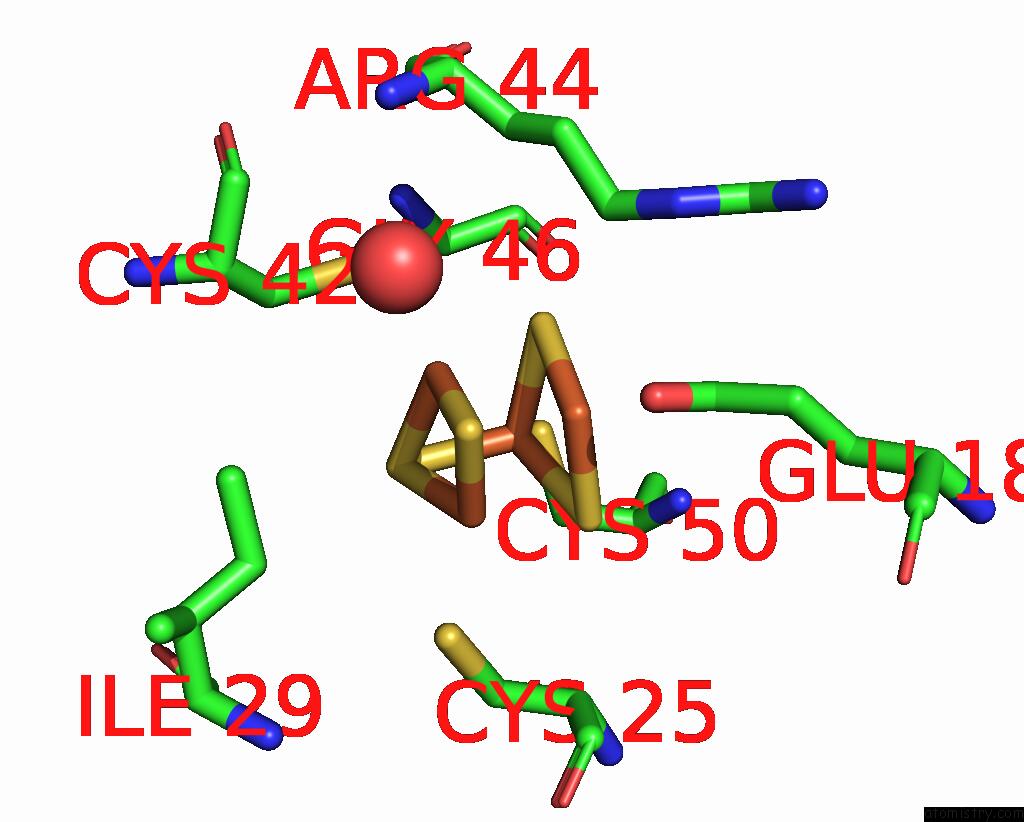
Mono view
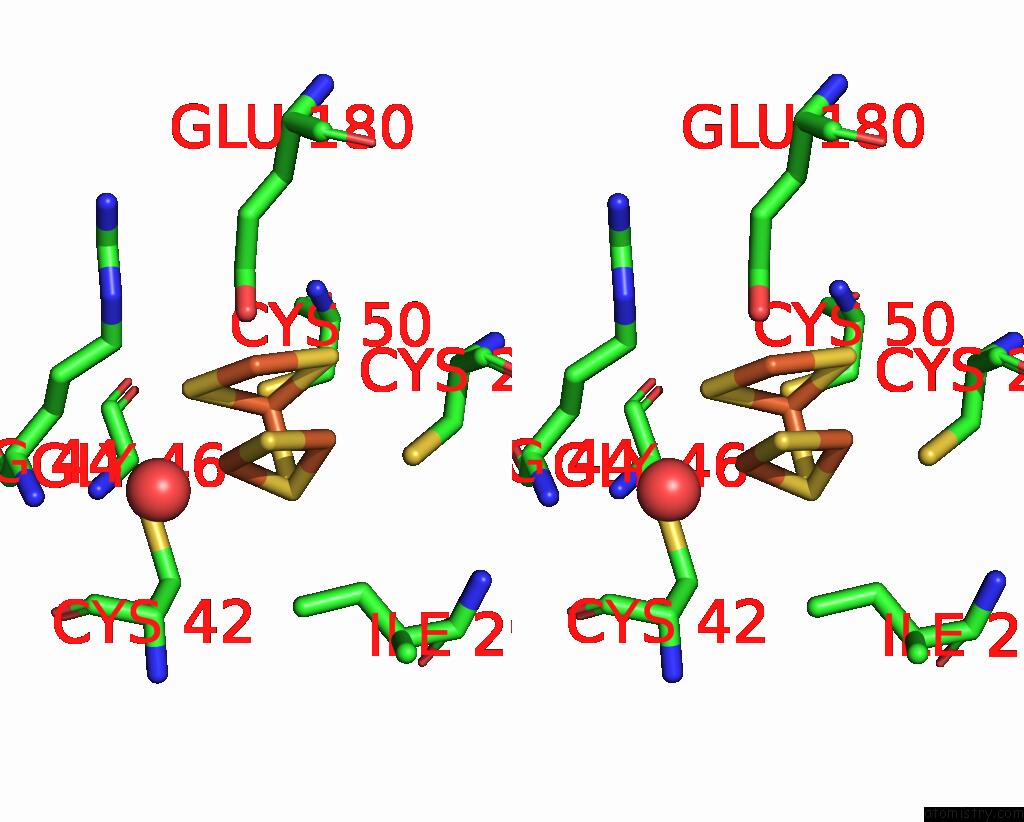
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 7 of Crystal Structure of the Coenzyme F420-Dependent Sulfite Reductase From Methanothermococcus Thermolithotrophicus at 1.55-A Resolution within 5.0Å range:
|
Iron binding site 8 out of 408 in 7npa
Go back to
Iron binding site 8 out
of 408 in the Crystal Structure of the Coenzyme F420-Dependent Sulfite Reductase From Methanothermococcus Thermolithotrophicus at 1.55-A Resolution
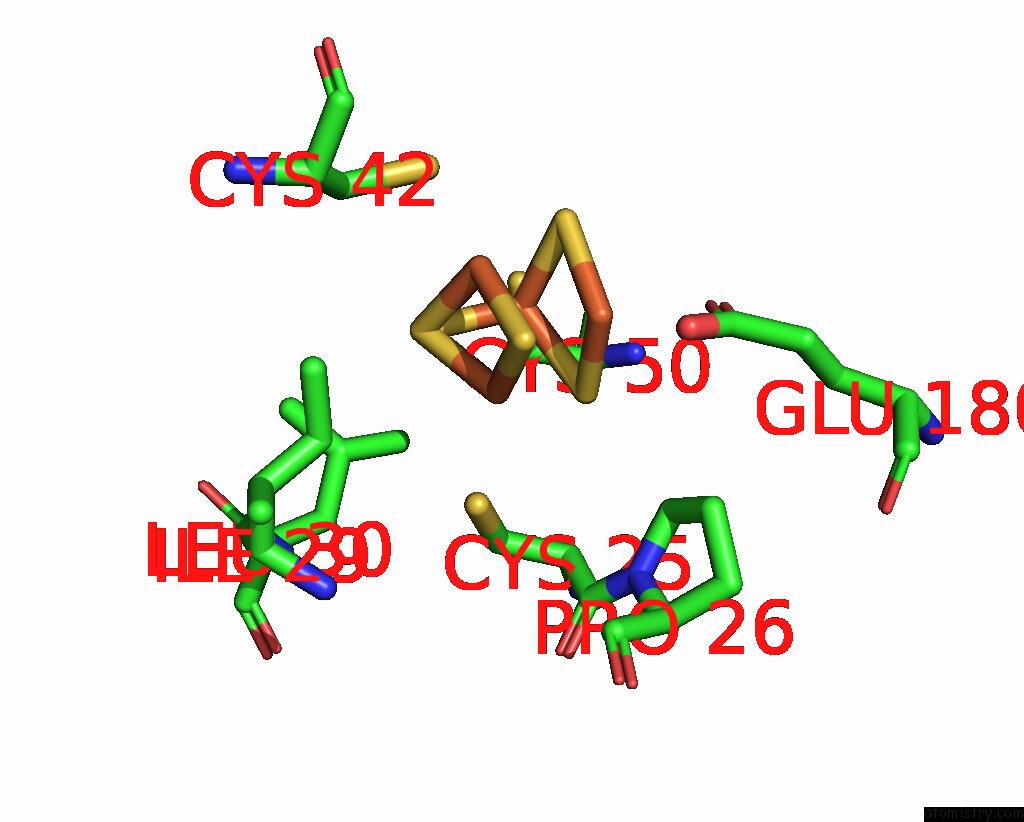
Mono view
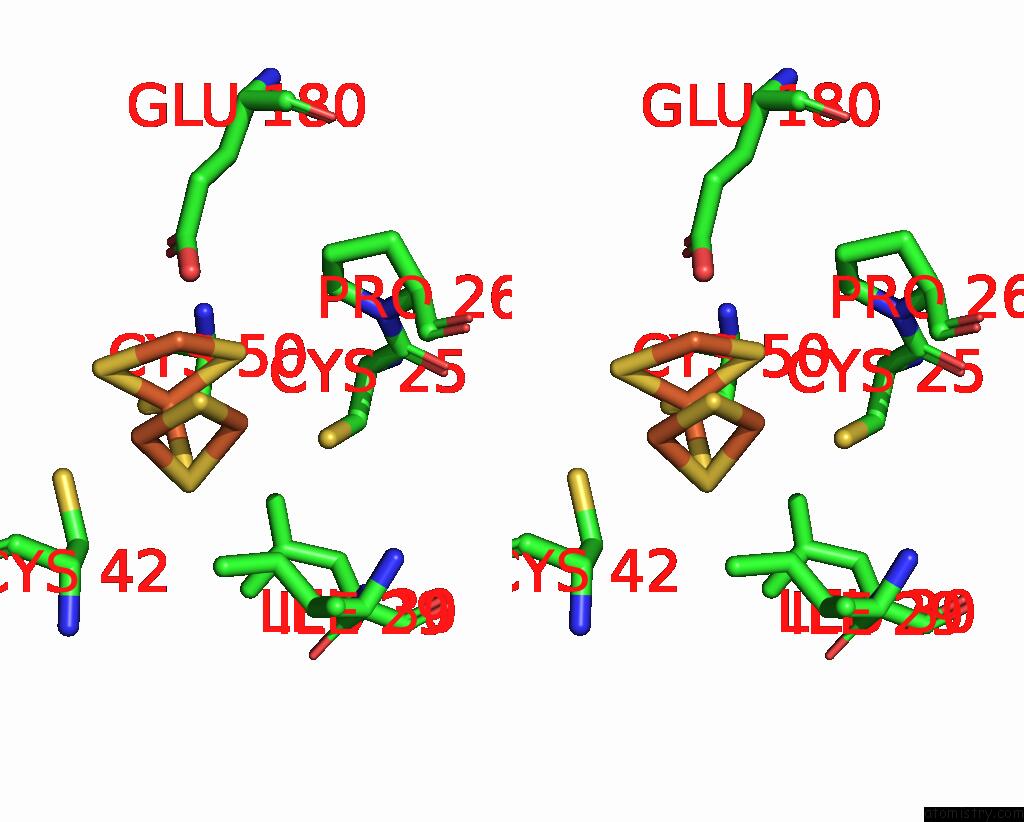
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 8 of Crystal Structure of the Coenzyme F420-Dependent Sulfite Reductase From Methanothermococcus Thermolithotrophicus at 1.55-A Resolution within 5.0Å range:
|
Iron binding site 9 out of 408 in 7npa
Go back to
Iron binding site 9 out
of 408 in the Crystal Structure of the Coenzyme F420-Dependent Sulfite Reductase From Methanothermococcus Thermolithotrophicus at 1.55-A Resolution
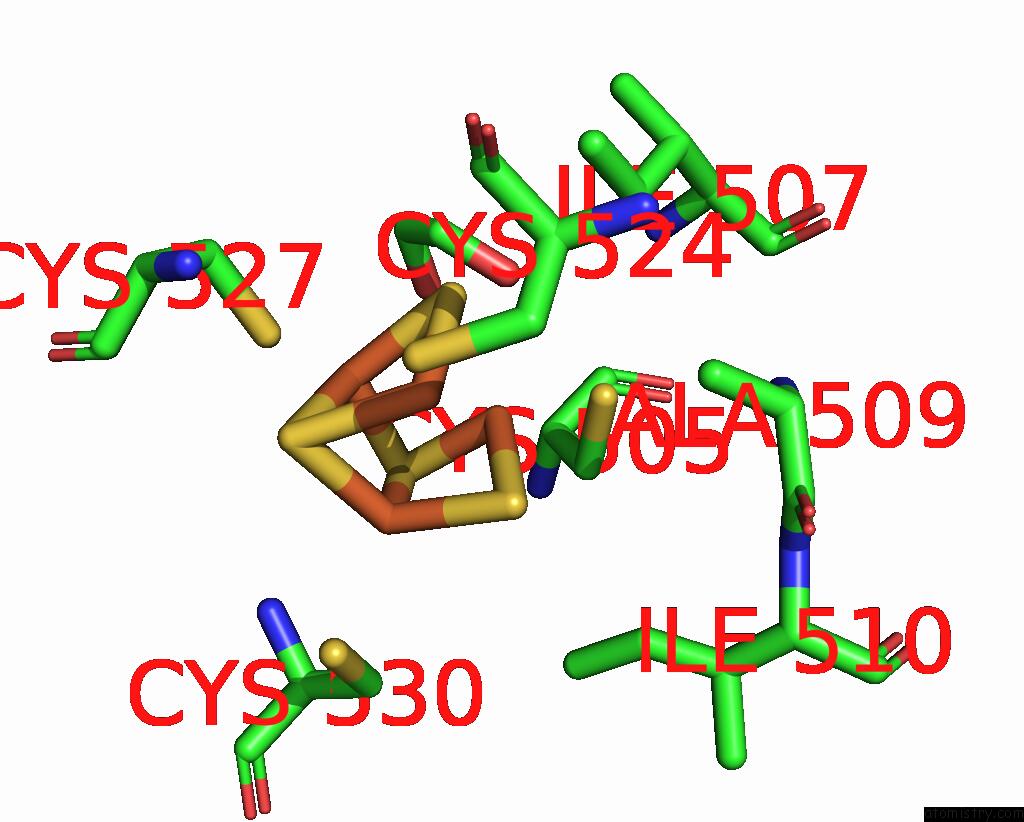
Mono view
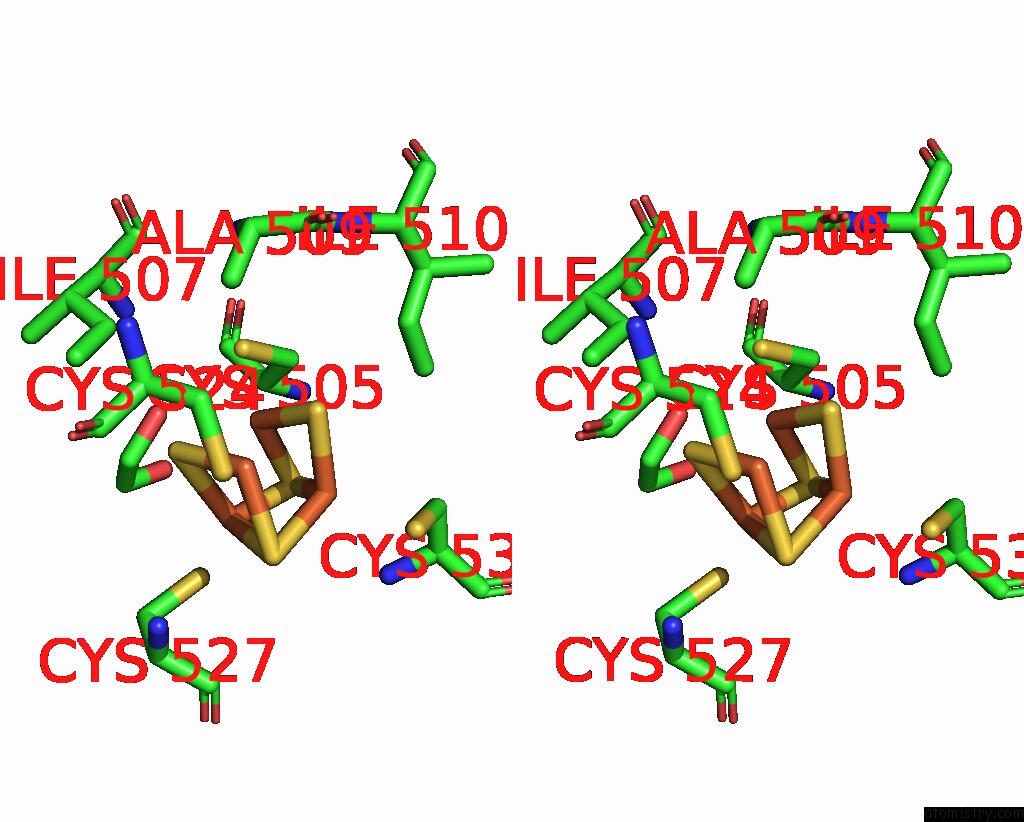
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 9 of Crystal Structure of the Coenzyme F420-Dependent Sulfite Reductase From Methanothermococcus Thermolithotrophicus at 1.55-A Resolution within 5.0Å range:
|
Iron binding site 10 out of 408 in 7npa
Go back to
Iron binding site 10 out
of 408 in the Crystal Structure of the Coenzyme F420-Dependent Sulfite Reductase From Methanothermococcus Thermolithotrophicus at 1.55-A Resolution
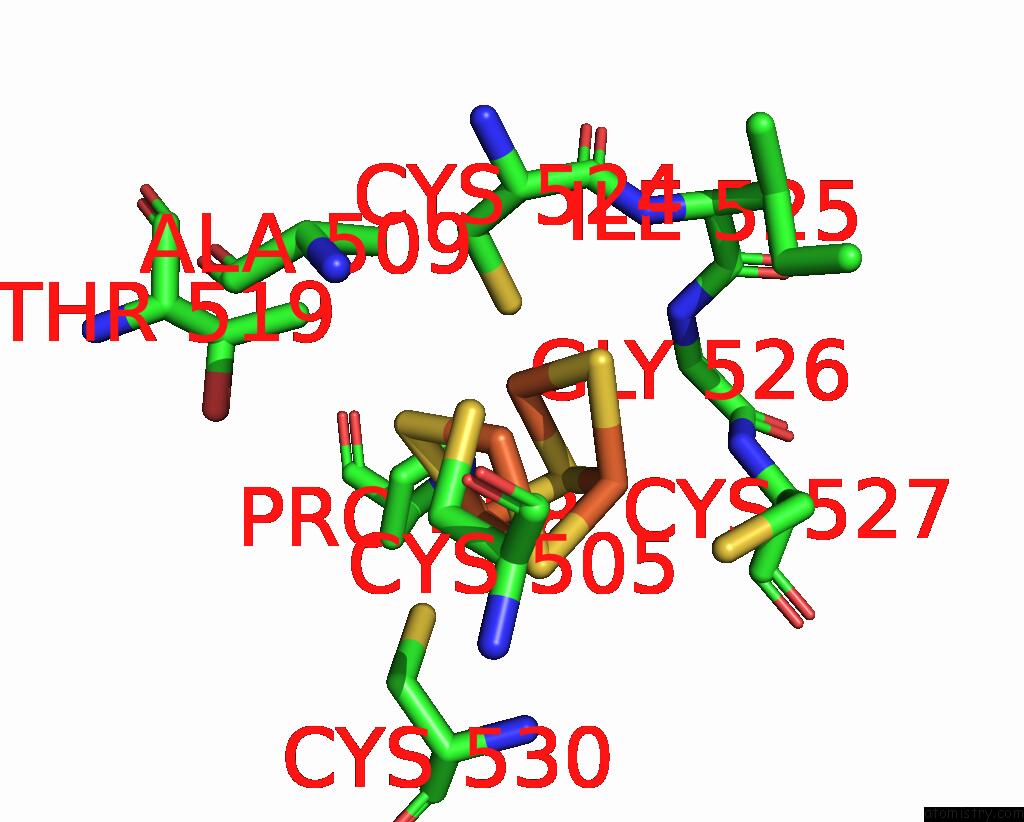
Mono view
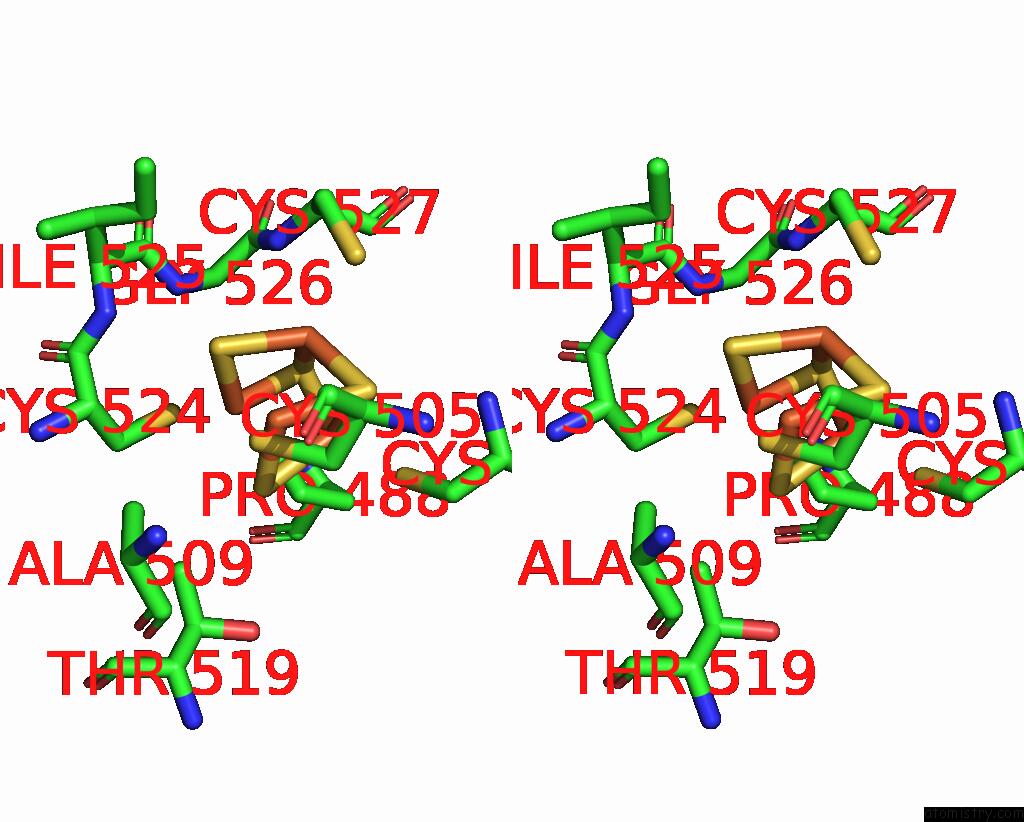
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 10 of Crystal Structure of the Coenzyme F420-Dependent Sulfite Reductase From Methanothermococcus Thermolithotrophicus at 1.55-A Resolution within 5.0Å range:
|
Reference:
M.Jespersen,
T.Wagner.
The Structure of the F420-Dependent Sulfite-Detoxifying Enzyme From Methanococcales Reveals A Prototypical Sulfite-Reductase with Assimilatory Traits. To Be Published.
Page generated: Thu Aug 8 09:50:40 2024
Last articles
F in 4P75F in 4P6G
F in 4P5Z
F in 4OZ3
F in 4P45
F in 4P1U
F in 4P44
F in 4P2H
F in 4OWO
F in 4OZ2