Iron »
PDB 7o4j-7oqr »
7opl »
Iron in PDB 7opl: Cryoem Structure of Dna Polymerase Alpha - Primase Bound to Sars Cov NSP1
Enzymatic activity of Cryoem Structure of Dna Polymerase Alpha - Primase Bound to Sars Cov NSP1
All present enzymatic activity of Cryoem Structure of Dna Polymerase Alpha - Primase Bound to Sars Cov NSP1:
2.7.7.7;
2.7.7.7;
Other elements in 7opl:
The structure of Cryoem Structure of Dna Polymerase Alpha - Primase Bound to Sars Cov NSP1 also contains other interesting chemical elements:
| Zinc | (Zn) | 3 atoms |
Iron Binding Sites:
The binding sites of Iron atom in the Cryoem Structure of Dna Polymerase Alpha - Primase Bound to Sars Cov NSP1
(pdb code 7opl). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 4 binding sites of Iron where determined in the Cryoem Structure of Dna Polymerase Alpha - Primase Bound to Sars Cov NSP1, PDB code: 7opl:
Jump to Iron binding site number: 1; 2; 3; 4;
In total 4 binding sites of Iron where determined in the Cryoem Structure of Dna Polymerase Alpha - Primase Bound to Sars Cov NSP1, PDB code: 7opl:
Jump to Iron binding site number: 1; 2; 3; 4;
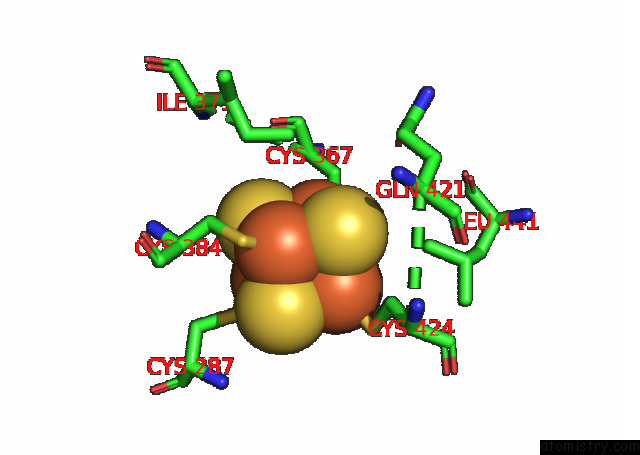
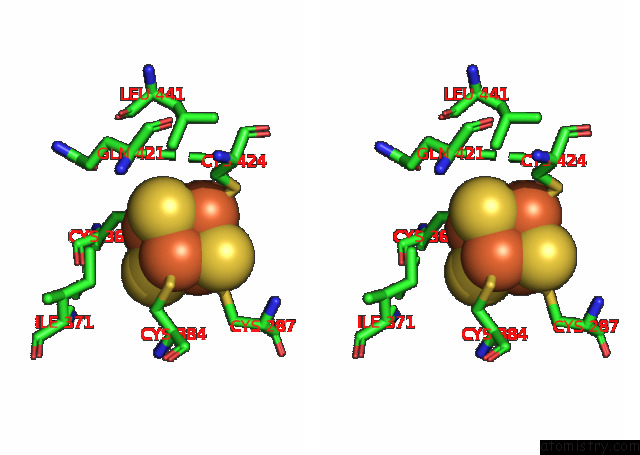
Iron binding site 1 out of 4 in 7opl
Go back to
Iron binding site 1 out
of 4 in the Cryoem Structure of Dna Polymerase Alpha - Primase Bound to Sars Cov NSP1
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Cryoem Structure of Dna Polymerase Alpha - Primase Bound to Sars Cov NSP1 within 5.0Å range:
|
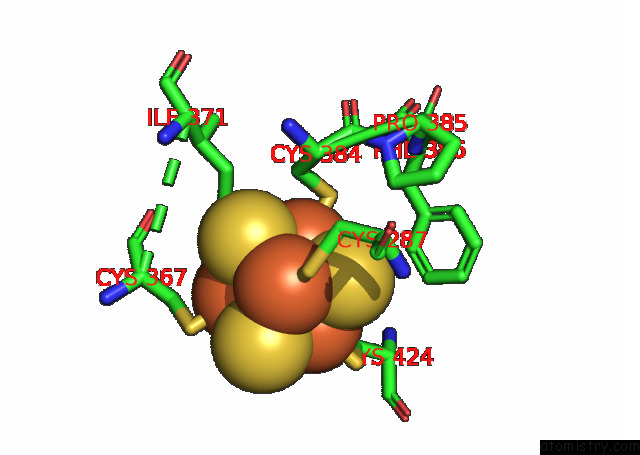
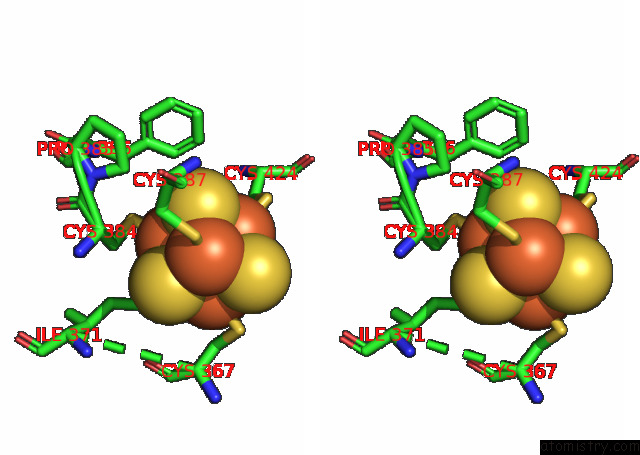
Iron binding site 2 out of 4 in 7opl
Go back to
Iron binding site 2 out
of 4 in the Cryoem Structure of Dna Polymerase Alpha - Primase Bound to Sars Cov NSP1
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Cryoem Structure of Dna Polymerase Alpha - Primase Bound to Sars Cov NSP1 within 5.0Å range:
|
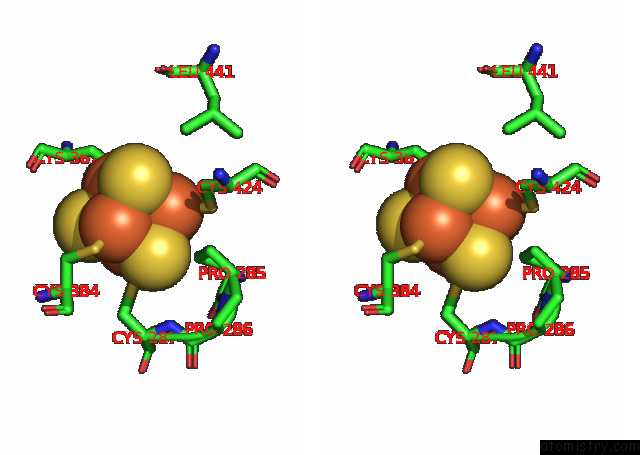
Iron binding site 3 out of 4 in 7opl
Go back to
Iron binding site 3 out
of 4 in the Cryoem Structure of Dna Polymerase Alpha - Primase Bound to Sars Cov NSP1
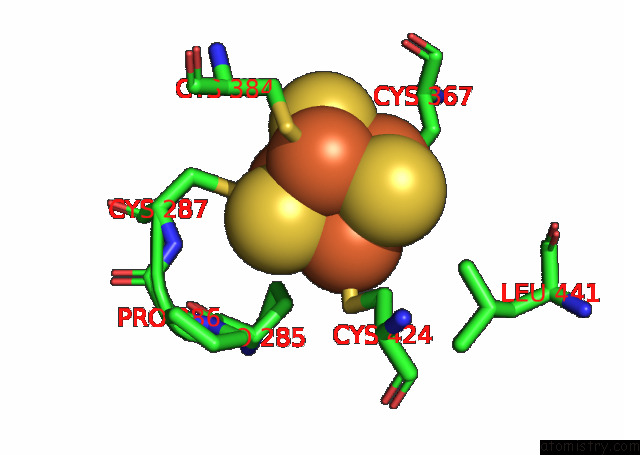
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 3 of Cryoem Structure of Dna Polymerase Alpha - Primase Bound to Sars Cov NSP1 within 5.0Å range:
|
Iron binding site 4 out of 4 in 7opl
Go back to
Iron binding site 4 out
of 4 in the Cryoem Structure of Dna Polymerase Alpha - Primase Bound to Sars Cov NSP1
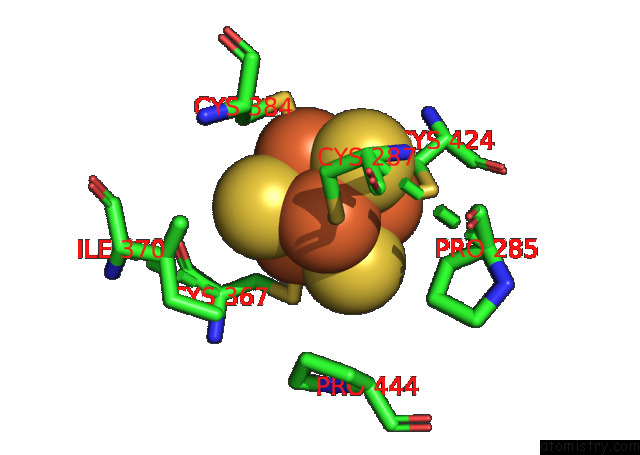
Mono view
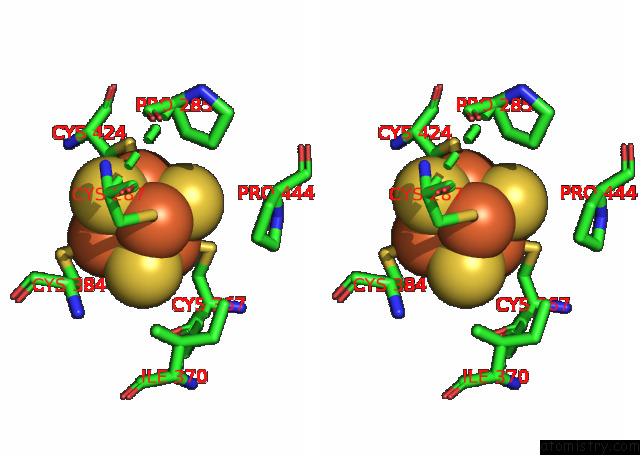
Stereo pair view

Mono view
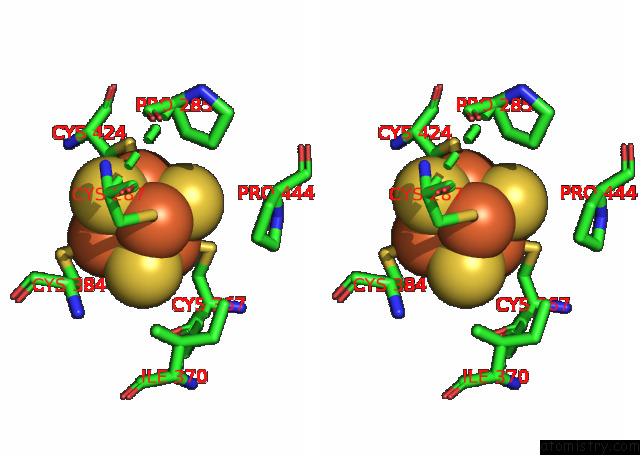
Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 4 of Cryoem Structure of Dna Polymerase Alpha - Primase Bound to Sars Cov NSP1 within 5.0Å range:
|
Reference:
M.L.Kilkenny,
C.E.Veale,
A.Guppy,
S.W.Hardwick,
D.Y.Chirgadze,
N.J.Rzechorzek,
J.D.Maman,
L.Pellegrini.
Structural Basis For the Interaction of Sars-Cov-2 Virulence Factor NSP1 with Dna Polymerase A - Primase. Protein Sci. 2021.
ISSN: ESSN 1469-896X
PubMed: 34719824
DOI: 10.1002/PRO.4220
Page generated: Thu Aug 8 14:27:14 2024
ISSN: ESSN 1469-896X
PubMed: 34719824
DOI: 10.1002/PRO.4220
Last articles
Cl in 5UQSCl in 5UQP
Cl in 5UQF
Cl in 5UPZ
Cl in 5UPR
Cl in 5UQA
Cl in 5UPQ
Cl in 5UPT
Cl in 5UPY
Cl in 5UPG