Iron »
PDB 7p7l-7pr2 »
7pim »
Iron in PDB 7pim: Partial Structure of Tyrosine Hydroxylase Lacking the First 35 Residues in Complex with Dopamine.
Enzymatic activity of Partial Structure of Tyrosine Hydroxylase Lacking the First 35 Residues in Complex with Dopamine.
All present enzymatic activity of Partial Structure of Tyrosine Hydroxylase Lacking the First 35 Residues in Complex with Dopamine.:
1.14.16.2;
1.14.16.2;
Iron Binding Sites:
The binding sites of Iron atom in the Partial Structure of Tyrosine Hydroxylase Lacking the First 35 Residues in Complex with Dopamine.
(pdb code 7pim). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 4 binding sites of Iron where determined in the Partial Structure of Tyrosine Hydroxylase Lacking the First 35 Residues in Complex with Dopamine., PDB code: 7pim:
Jump to Iron binding site number: 1; 2; 3; 4;
In total 4 binding sites of Iron where determined in the Partial Structure of Tyrosine Hydroxylase Lacking the First 35 Residues in Complex with Dopamine., PDB code: 7pim:
Jump to Iron binding site number: 1; 2; 3; 4;
Iron binding site 1 out of 4 in 7pim
Go back to
Iron binding site 1 out
of 4 in the Partial Structure of Tyrosine Hydroxylase Lacking the First 35 Residues in Complex with Dopamine.
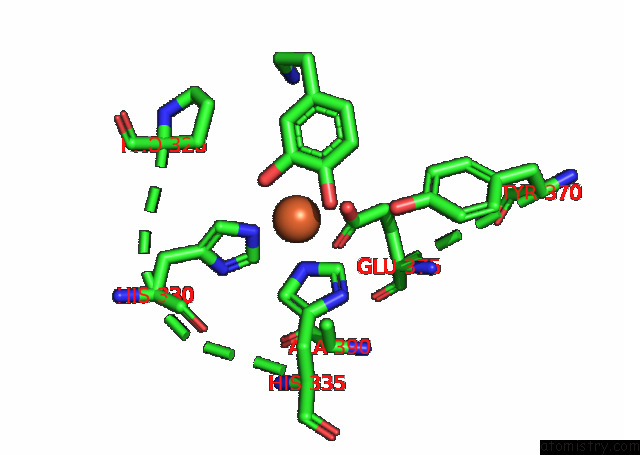
Mono view
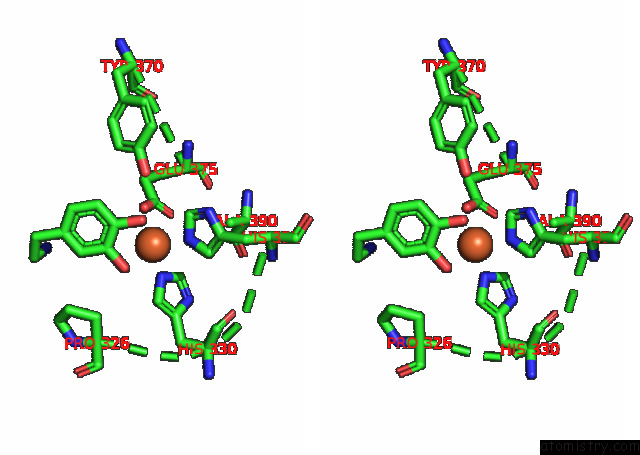
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Partial Structure of Tyrosine Hydroxylase Lacking the First 35 Residues in Complex with Dopamine. within 5.0Å range:
|
Iron binding site 2 out of 4 in 7pim
Go back to
Iron binding site 2 out
of 4 in the Partial Structure of Tyrosine Hydroxylase Lacking the First 35 Residues in Complex with Dopamine.
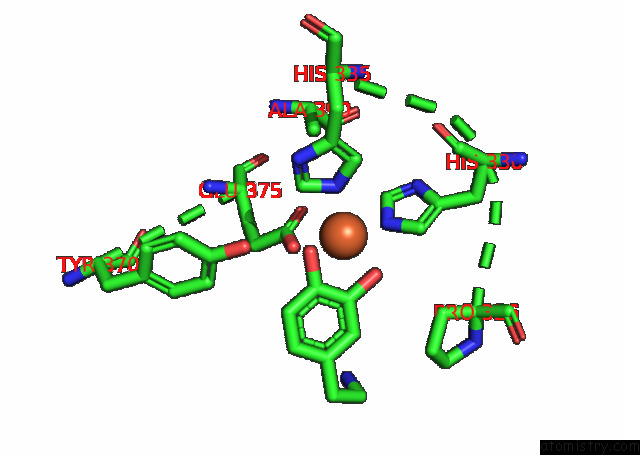
Mono view
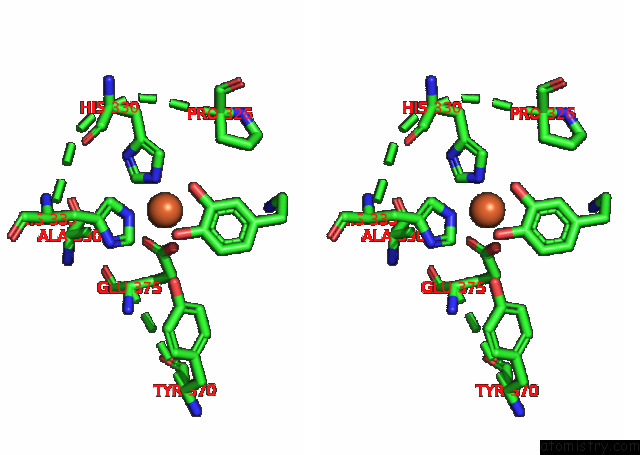
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Partial Structure of Tyrosine Hydroxylase Lacking the First 35 Residues in Complex with Dopamine. within 5.0Å range:
|
Iron binding site 3 out of 4 in 7pim
Go back to
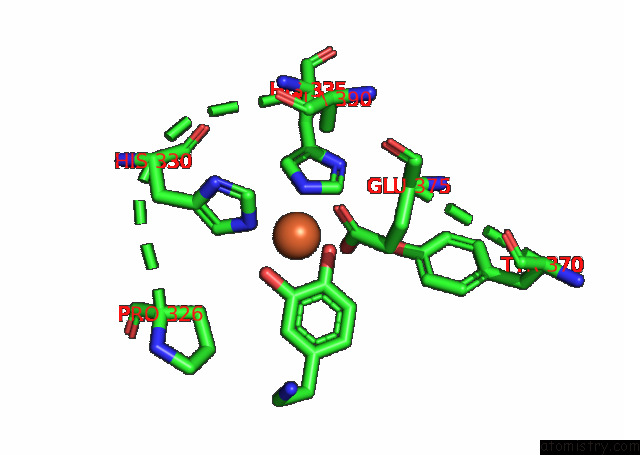
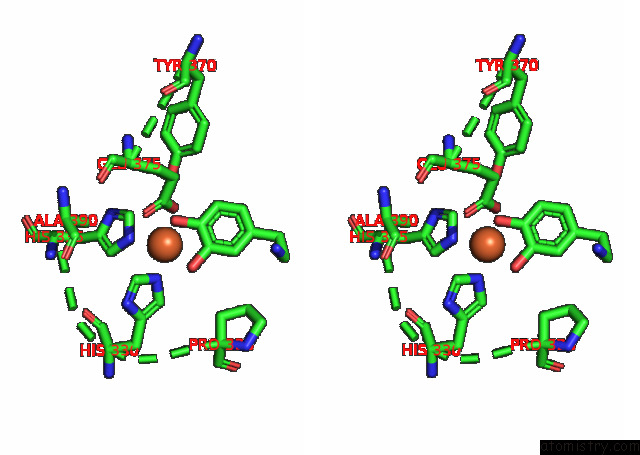
Iron binding site 3 out
of 4 in the Partial Structure of Tyrosine Hydroxylase Lacking the First 35 Residues in Complex with Dopamine.
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 3 of Partial Structure of Tyrosine Hydroxylase Lacking the First 35 Residues in Complex with Dopamine. within 5.0Å range:
|
Iron binding site 4 out of 4 in 7pim
Go back to
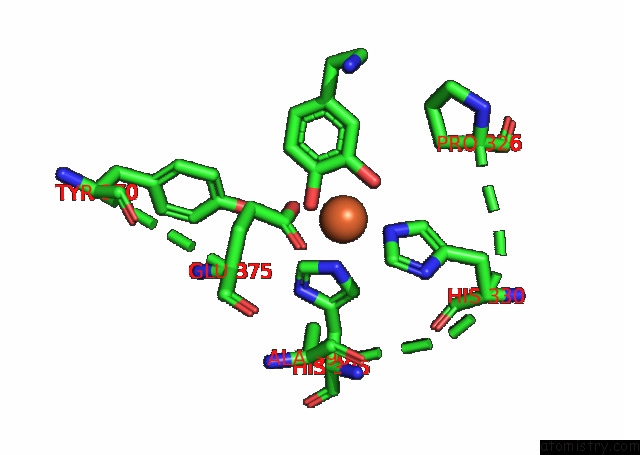
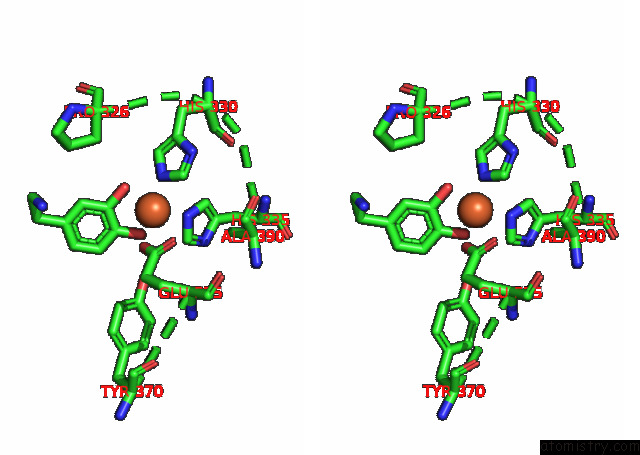
Iron binding site 4 out
of 4 in the Partial Structure of Tyrosine Hydroxylase Lacking the First 35 Residues in Complex with Dopamine.
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 4 of Partial Structure of Tyrosine Hydroxylase Lacking the First 35 Residues in Complex with Dopamine. within 5.0Å range:
|
Reference:
M.T.Bueno-Carrasco,
J.Cuellar,
M.I.Flydal,
C.Santiago,
T.A.Krakenes,
R.Kleppe,
J.R.Lopez-Blanco,
K.Teigen,
S.Alvira,
P.Chacon,
A.Martinez,
J.M.Valpuesta.
Structural Mechanism For Tyrosine Hydroxylase Inhibition By Dopamine and Reactivation By SER40 Phosphorylation To Be Published.
Page generated: Thu Aug 7 02:52:20 2025
Last articles
Mg in 1RTDMg in 1RTK
Mg in 1RRP
Mg in 1RS0
Mg in 1RRG
Mg in 1RRF
Mg in 1RQJ
Mg in 1RQY
Mg in 1RQN
Mg in 1RQI