Iron »
PDB 7rkw-7scp »
7s6r »
Iron in PDB 7s6r: Complex Structure of Methane Monooxygenase Hydroxylase and Regulatory Subunit with H5A Mutation
Protein crystallography data
The structure of Complex Structure of Methane Monooxygenase Hydroxylase and Regulatory Subunit with H5A Mutation, PDB code: 7s6r
was solved by
J.C.Johns,
R.Banerjee,
M.M.Semonis,
K.Shi,
H.Aihara,
J.D.Lipscomb,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 73.71 / 1.89 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 102.655, 105.907, 299.12, 90, 90, 90 |
| R / Rfree (%) | 16.1 / 19.5 |
Iron Binding Sites:
The binding sites of Iron atom in the Complex Structure of Methane Monooxygenase Hydroxylase and Regulatory Subunit with H5A Mutation
(pdb code 7s6r). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 4 binding sites of Iron where determined in the Complex Structure of Methane Monooxygenase Hydroxylase and Regulatory Subunit with H5A Mutation, PDB code: 7s6r:
Jump to Iron binding site number: 1; 2; 3; 4;
In total 4 binding sites of Iron where determined in the Complex Structure of Methane Monooxygenase Hydroxylase and Regulatory Subunit with H5A Mutation, PDB code: 7s6r:
Jump to Iron binding site number: 1; 2; 3; 4;
Iron binding site 1 out of 4 in 7s6r
Go back to
Iron binding site 1 out
of 4 in the Complex Structure of Methane Monooxygenase Hydroxylase and Regulatory Subunit with H5A Mutation
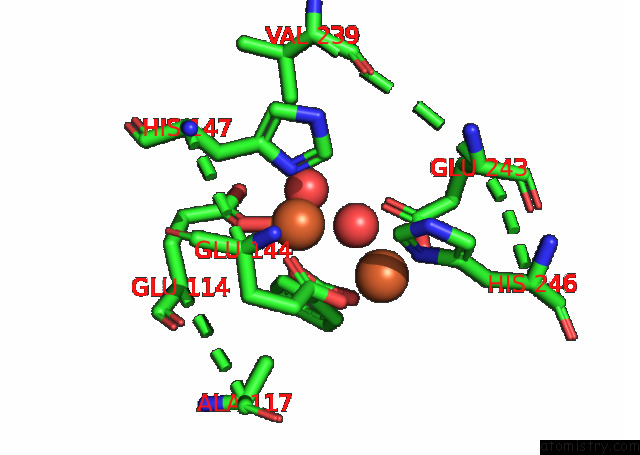
Mono view
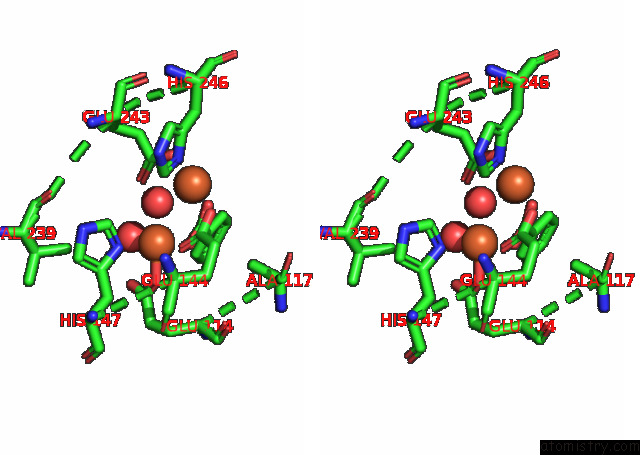
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Complex Structure of Methane Monooxygenase Hydroxylase and Regulatory Subunit with H5A Mutation within 5.0Å range:
|
Iron binding site 2 out of 4 in 7s6r
Go back to
Iron binding site 2 out
of 4 in the Complex Structure of Methane Monooxygenase Hydroxylase and Regulatory Subunit with H5A Mutation
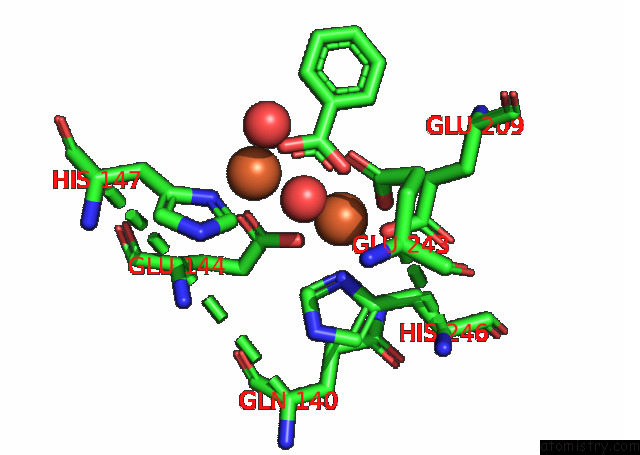
Mono view
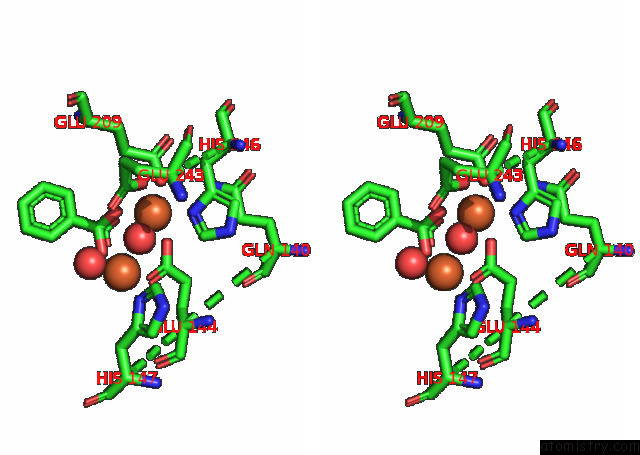
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Complex Structure of Methane Monooxygenase Hydroxylase and Regulatory Subunit with H5A Mutation within 5.0Å range:
|
Iron binding site 3 out of 4 in 7s6r
Go back to
Iron binding site 3 out
of 4 in the Complex Structure of Methane Monooxygenase Hydroxylase and Regulatory Subunit with H5A Mutation
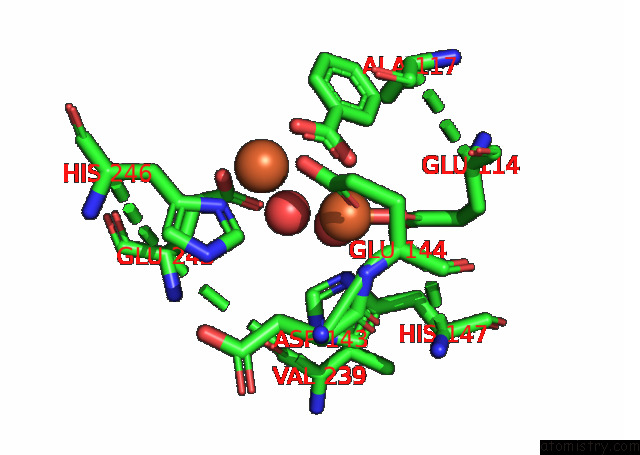
Mono view
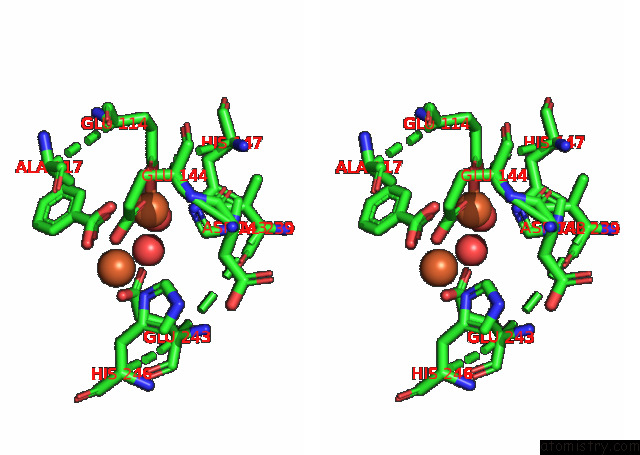
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 3 of Complex Structure of Methane Monooxygenase Hydroxylase and Regulatory Subunit with H5A Mutation within 5.0Å range:
|
Iron binding site 4 out of 4 in 7s6r
Go back to
Iron binding site 4 out
of 4 in the Complex Structure of Methane Monooxygenase Hydroxylase and Regulatory Subunit with H5A Mutation
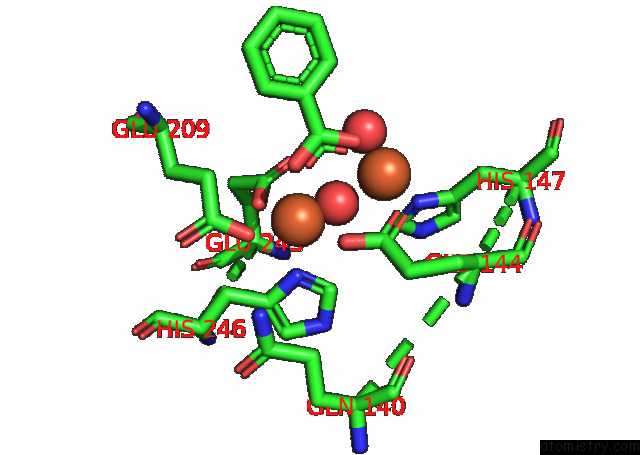
Mono view
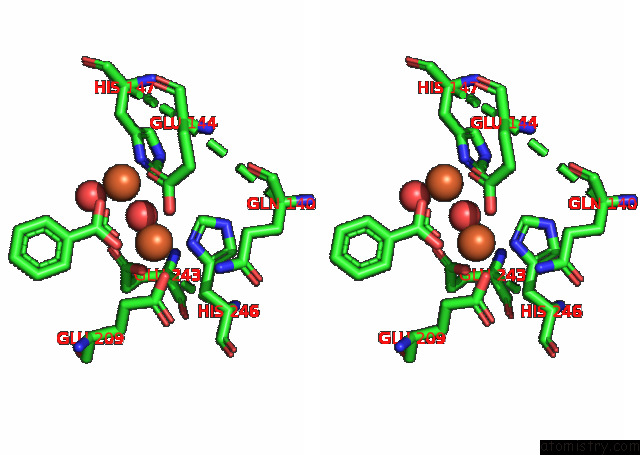
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 4 of Complex Structure of Methane Monooxygenase Hydroxylase and Regulatory Subunit with H5A Mutation within 5.0Å range:
|
Reference:
J.C.Jones,
R.Banerjee,
M.M.Semonis,
K.Shi,
H.Aihara,
J.D.Lipscomb.
X-Ray Crystal Structures of Methane Monooxygenase Hydroxylase Complexes with Variants of Its Regulatory Component: Correlations with Altered Reaction Cycle Dynamics. Biochemistry 2021.
ISSN: ISSN 0006-2960
PubMed: 34910460
DOI: 10.1021/ACS.BIOCHEM.1C00673
Page generated: Fri Aug 9 00:22:14 2024
ISSN: ISSN 0006-2960
PubMed: 34910460
DOI: 10.1021/ACS.BIOCHEM.1C00673
Last articles
Ca in 5TA5Ca in 5TAC
Ca in 5TA0
Ca in 5TA1
Ca in 5T9Q
Ca in 5T9K
Ca in 5T9I
Ca in 5T8D
Ca in 5T9C
Ca in 5T9B