Iron »
PDB 7tce-7tqh »
7tl4 »
Iron in PDB 7tl4: Crystal Structure of Yeast P58C Multi-Tyrosine Mutant 6YF
Protein crystallography data
The structure of Crystal Structure of Yeast P58C Multi-Tyrosine Mutant 6YF, PDB code: 7tl4
was solved by
A.M.Blee,
L.E.Salay,
W.J.Chazin,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.61 / 1.81 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 39.519, 51.488, 90.027, 90, 90, 90 |
| R / Rfree (%) | 19.6 / 21.4 |
Iron Binding Sites:
The binding sites of Iron atom in the Crystal Structure of Yeast P58C Multi-Tyrosine Mutant 6YF
(pdb code 7tl4). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 4 binding sites of Iron where determined in the Crystal Structure of Yeast P58C Multi-Tyrosine Mutant 6YF, PDB code: 7tl4:
Jump to Iron binding site number: 1; 2; 3; 4;
In total 4 binding sites of Iron where determined in the Crystal Structure of Yeast P58C Multi-Tyrosine Mutant 6YF, PDB code: 7tl4:
Jump to Iron binding site number: 1; 2; 3; 4;
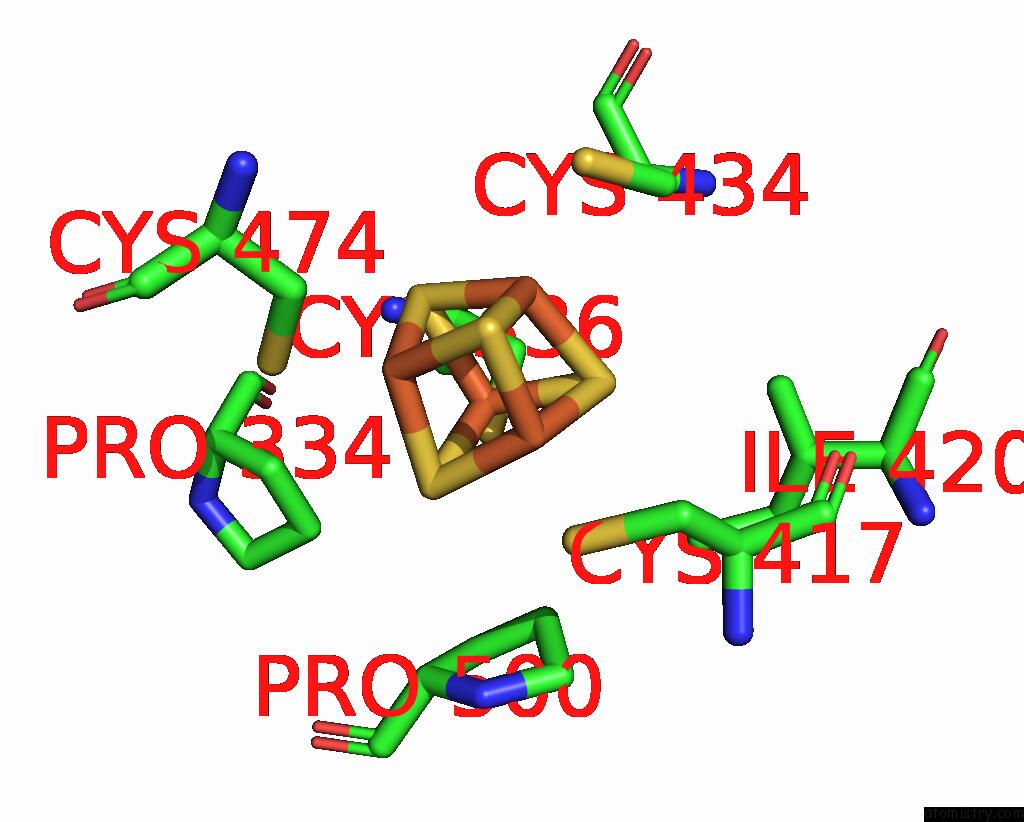
Iron binding site 1 out of 4 in 7tl4
Go back to
Iron binding site 1 out
of 4 in the Crystal Structure of Yeast P58C Multi-Tyrosine Mutant 6YF
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Crystal Structure of Yeast P58C Multi-Tyrosine Mutant 6YF within 5.0Å range:
|
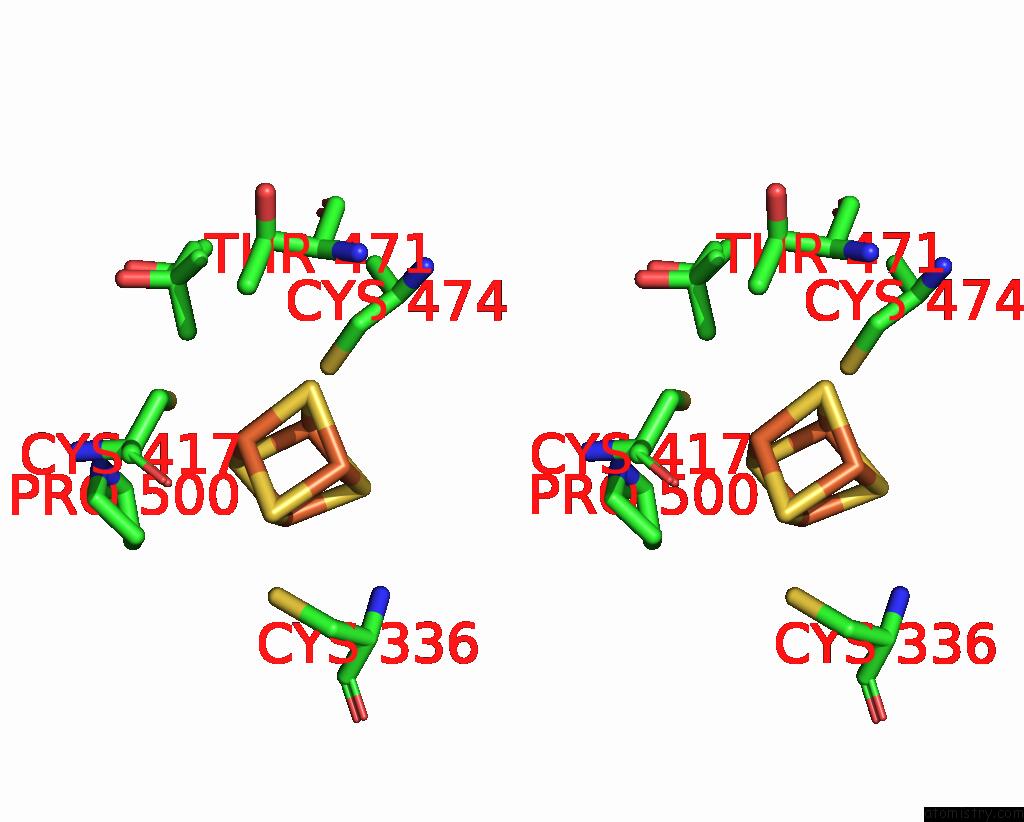
Iron binding site 2 out of 4 in 7tl4
Go back to
Iron binding site 2 out
of 4 in the Crystal Structure of Yeast P58C Multi-Tyrosine Mutant 6YF

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Crystal Structure of Yeast P58C Multi-Tyrosine Mutant 6YF within 5.0Å range:
|
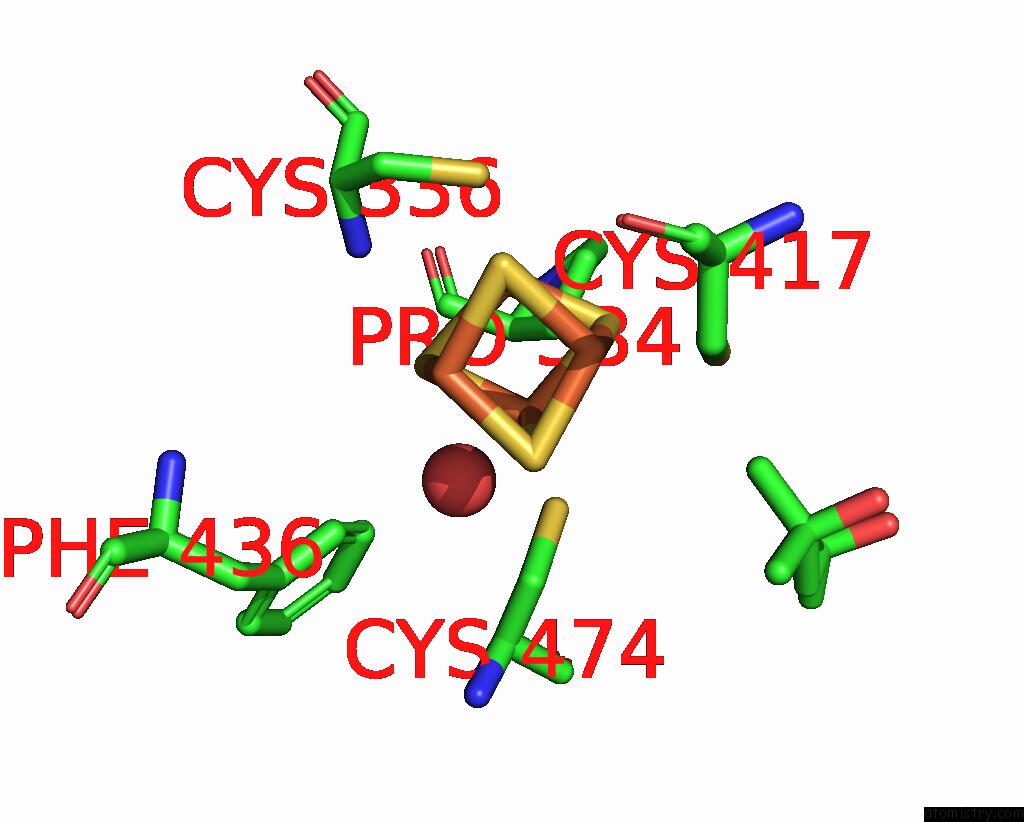
Iron binding site 3 out of 4 in 7tl4
Go back to
Iron binding site 3 out
of 4 in the Crystal Structure of Yeast P58C Multi-Tyrosine Mutant 6YF
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 3 of Crystal Structure of Yeast P58C Multi-Tyrosine Mutant 6YF within 5.0Å range:
|
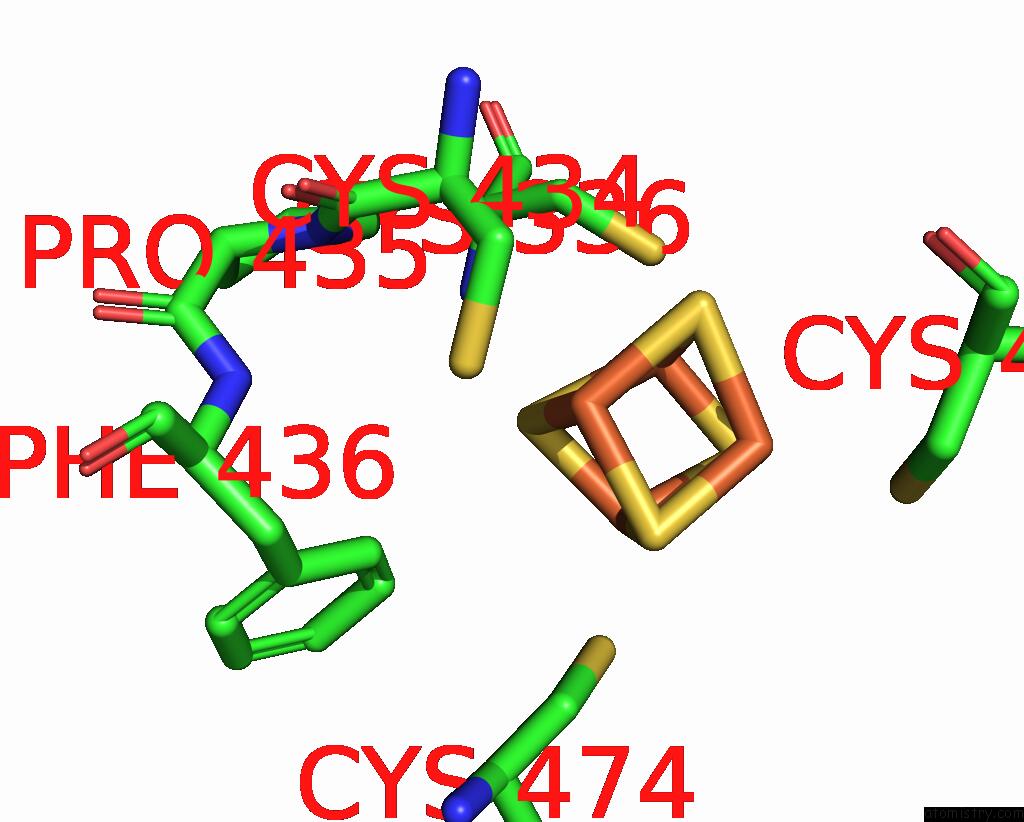
Iron binding site 4 out of 4 in 7tl4
Go back to
Iron binding site 4 out
of 4 in the Crystal Structure of Yeast P58C Multi-Tyrosine Mutant 6YF
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 4 of Crystal Structure of Yeast P58C Multi-Tyrosine Mutant 6YF within 5.0Å range:
|
Reference:
L.E.Salay,
A.M.Blee,
M.K.Raza,
K.S.Gallagher,
H.Chen,
A.J.Dorfeuille,
J.K.Barton,
W.J.Chazin.
Modification of the 4FE-4S Cluster Charge Transport Pathway Alters Rna Synthesis By Yeast Dna Primase. Biochemistry V. 61 1113 2022.
ISSN: ISSN 0006-2960
PubMed: 35617695
DOI: 10.1021/ACS.BIOCHEM.2C00100
Page generated: Thu Aug 7 05:57:39 2025
ISSN: ISSN 0006-2960
PubMed: 35617695
DOI: 10.1021/ACS.BIOCHEM.2C00100
Last articles
Mg in 1JWYMg in 1JUE
Mg in 1JUY
Mg in 1JUB
Mg in 1JSC
Mg in 1JS9
Mg in 1JR4
Mg in 1JPM
Mg in 1JQX
Mg in 1JQV