Iron »
PDB 8ire-8jfw »
8j5r »
Iron in PDB 8j5r: Cryo-Em Structure of Mycobacterium Tuberculosis Oppabcd in the Resting State
Iron Binding Sites:
The binding sites of Iron atom in the Cryo-Em Structure of Mycobacterium Tuberculosis Oppabcd in the Resting State
(pdb code 8j5r). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 4 binding sites of Iron where determined in the Cryo-Em Structure of Mycobacterium Tuberculosis Oppabcd in the Resting State, PDB code: 8j5r:
Jump to Iron binding site number: 1; 2; 3; 4;
In total 4 binding sites of Iron where determined in the Cryo-Em Structure of Mycobacterium Tuberculosis Oppabcd in the Resting State, PDB code: 8j5r:
Jump to Iron binding site number: 1; 2; 3; 4;
Iron binding site 1 out of 4 in 8j5r
Go back to
Iron binding site 1 out
of 4 in the Cryo-Em Structure of Mycobacterium Tuberculosis Oppabcd in the Resting State
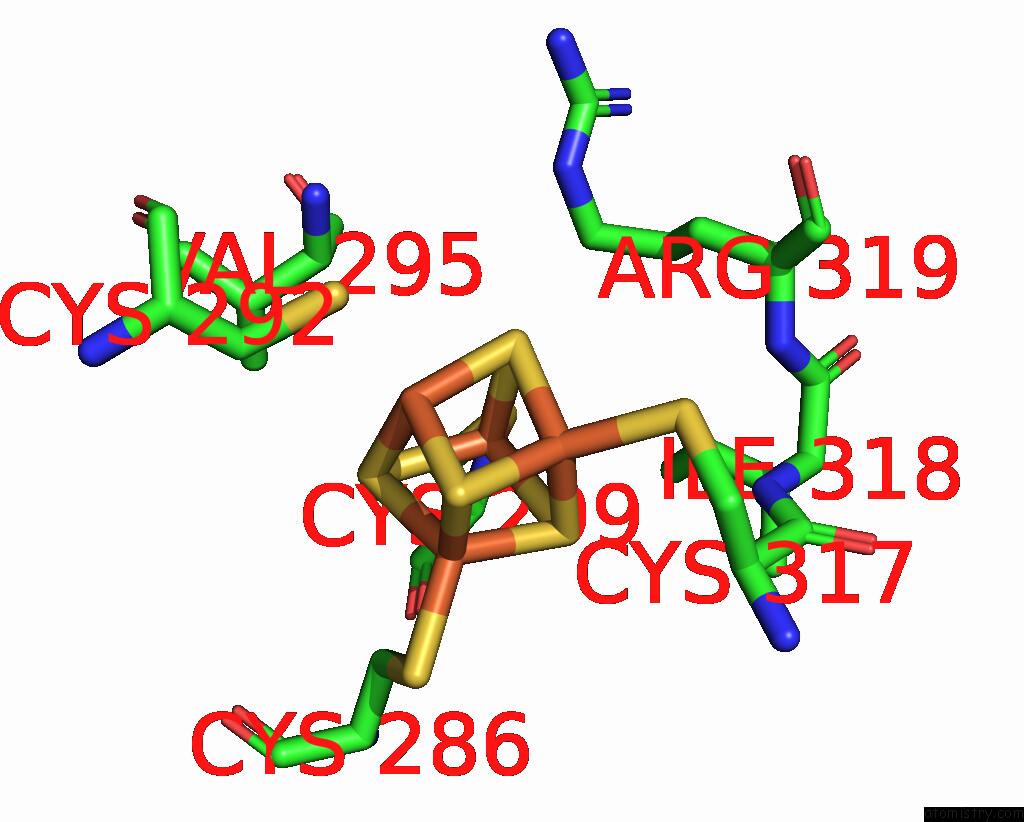
Mono view
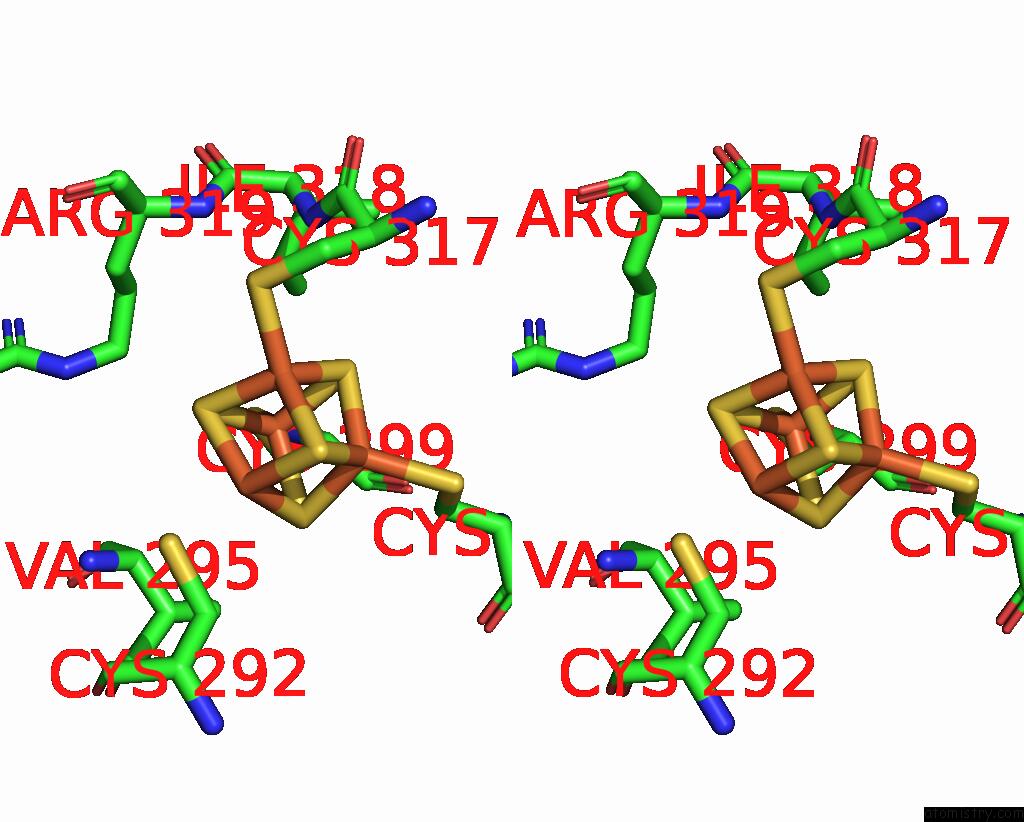
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Cryo-Em Structure of Mycobacterium Tuberculosis Oppabcd in the Resting State within 5.0Å range:
|
Iron binding site 2 out of 4 in 8j5r
Go back to
Iron binding site 2 out
of 4 in the Cryo-Em Structure of Mycobacterium Tuberculosis Oppabcd in the Resting State
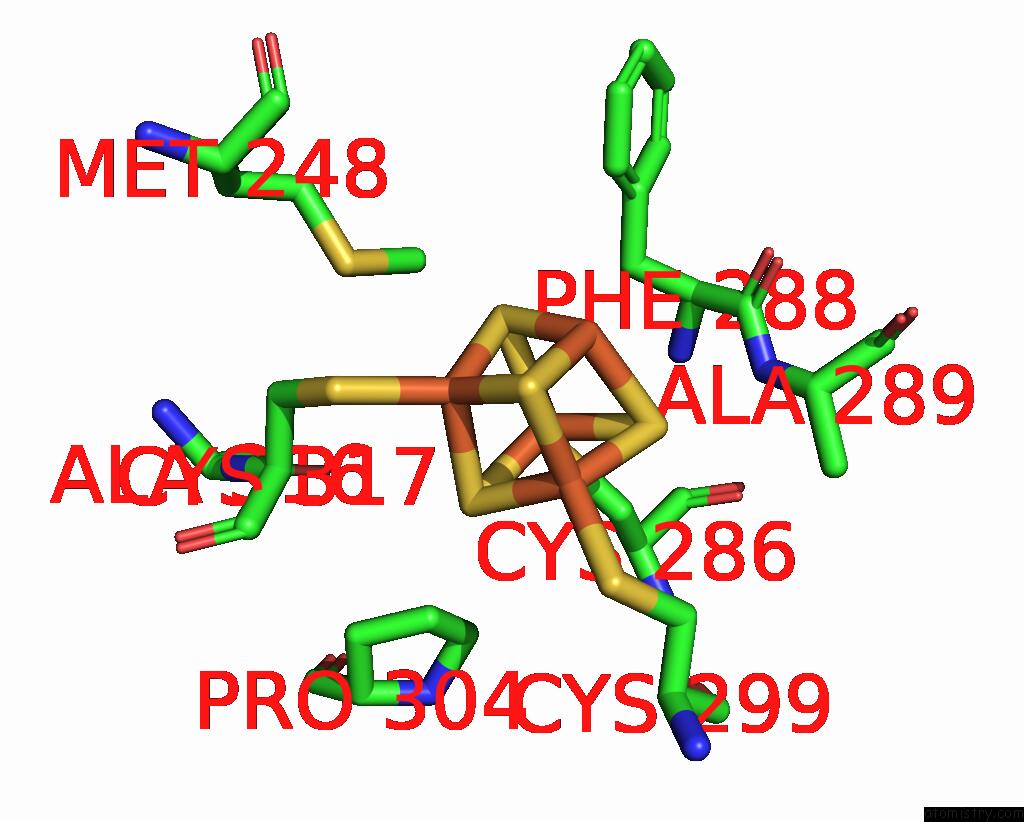
Mono view
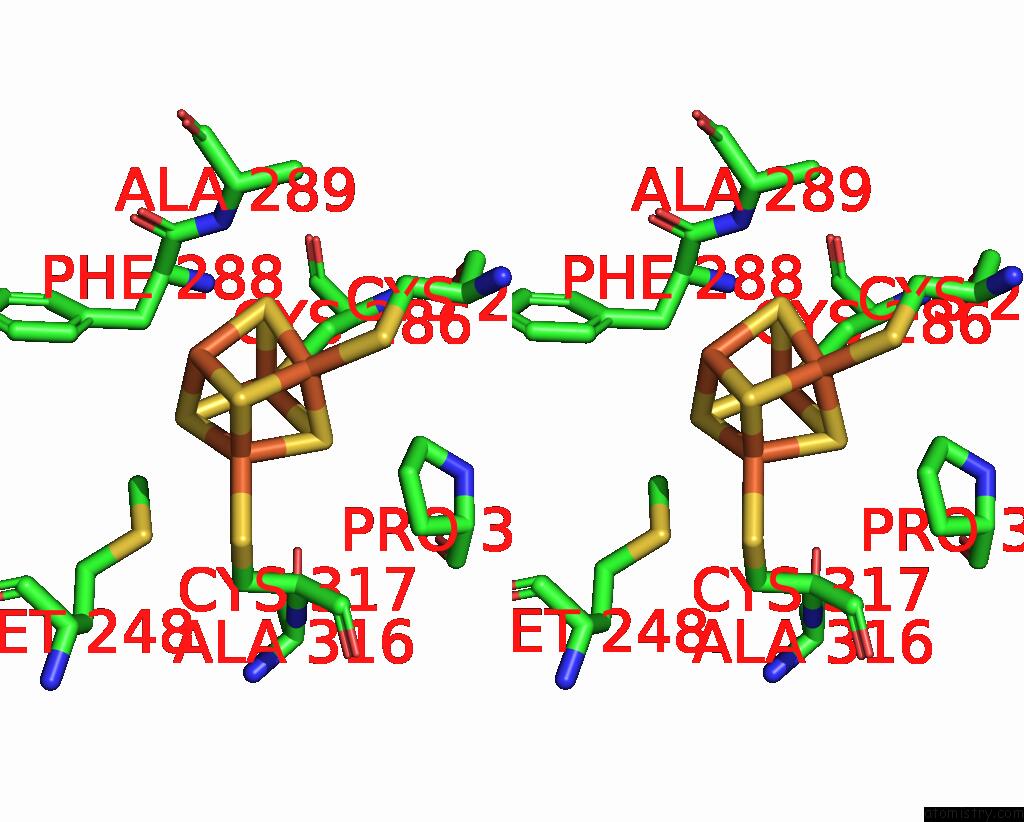
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Cryo-Em Structure of Mycobacterium Tuberculosis Oppabcd in the Resting State within 5.0Å range:
|
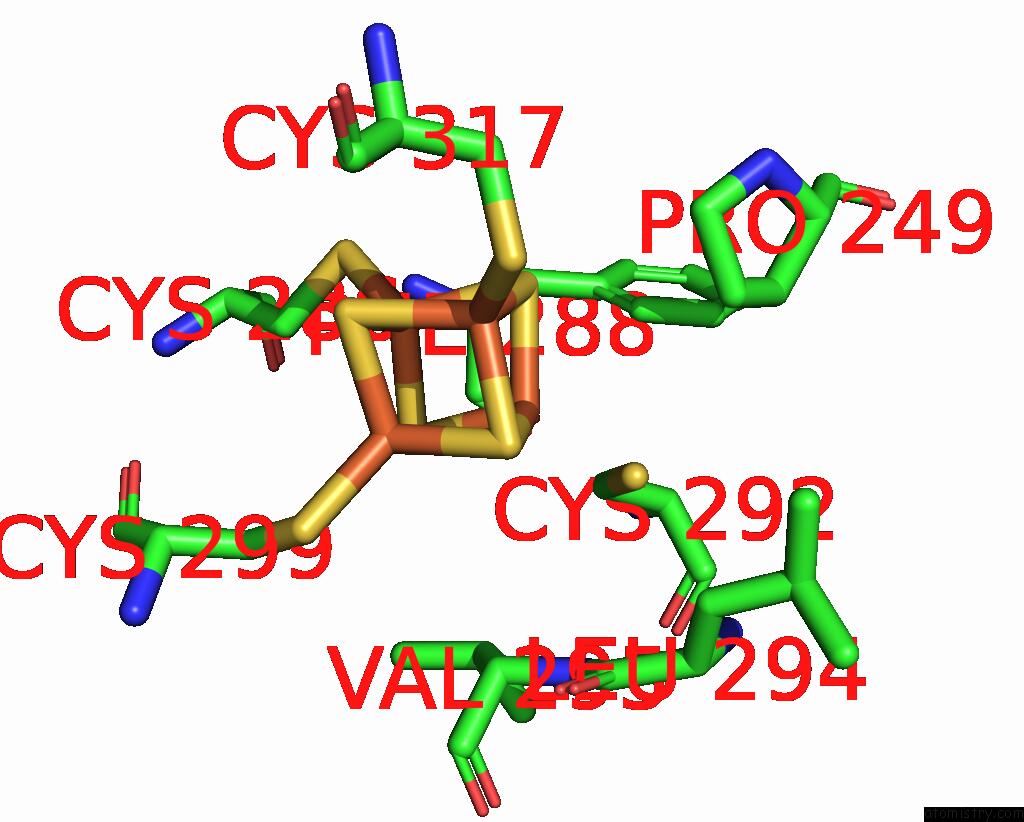
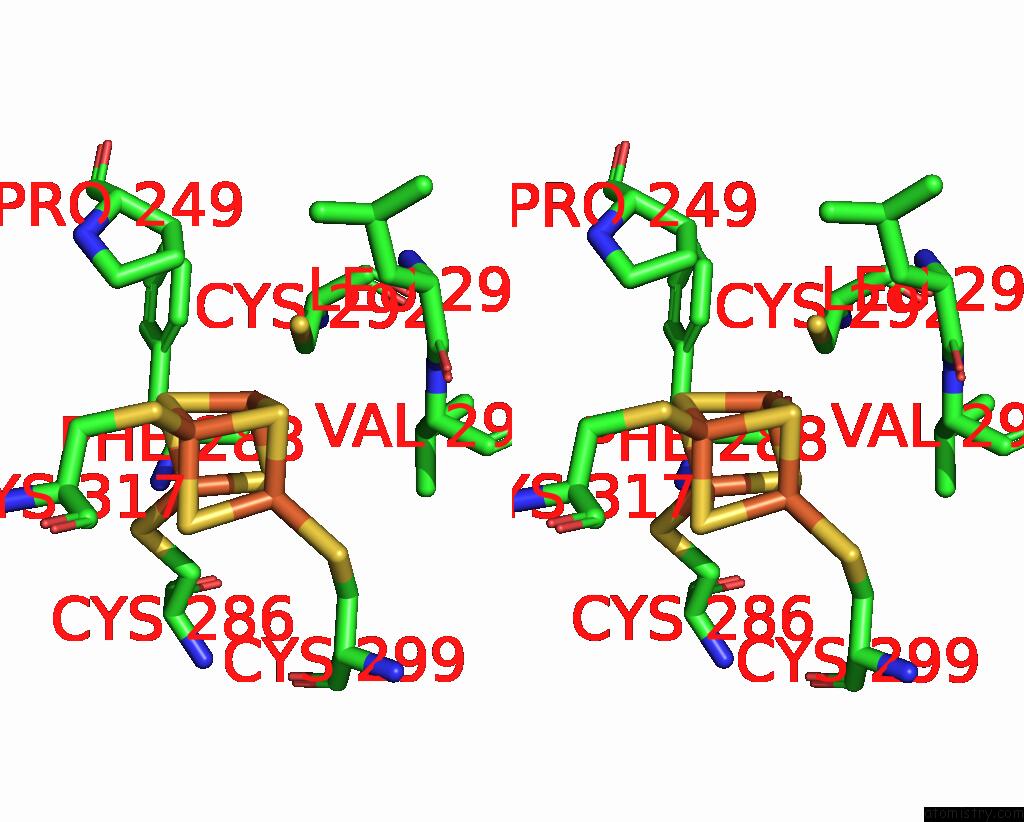
Iron binding site 3 out of 4 in 8j5r
Go back to
Iron binding site 3 out
of 4 in the Cryo-Em Structure of Mycobacterium Tuberculosis Oppabcd in the Resting State
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 3 of Cryo-Em Structure of Mycobacterium Tuberculosis Oppabcd in the Resting State within 5.0Å range:
|
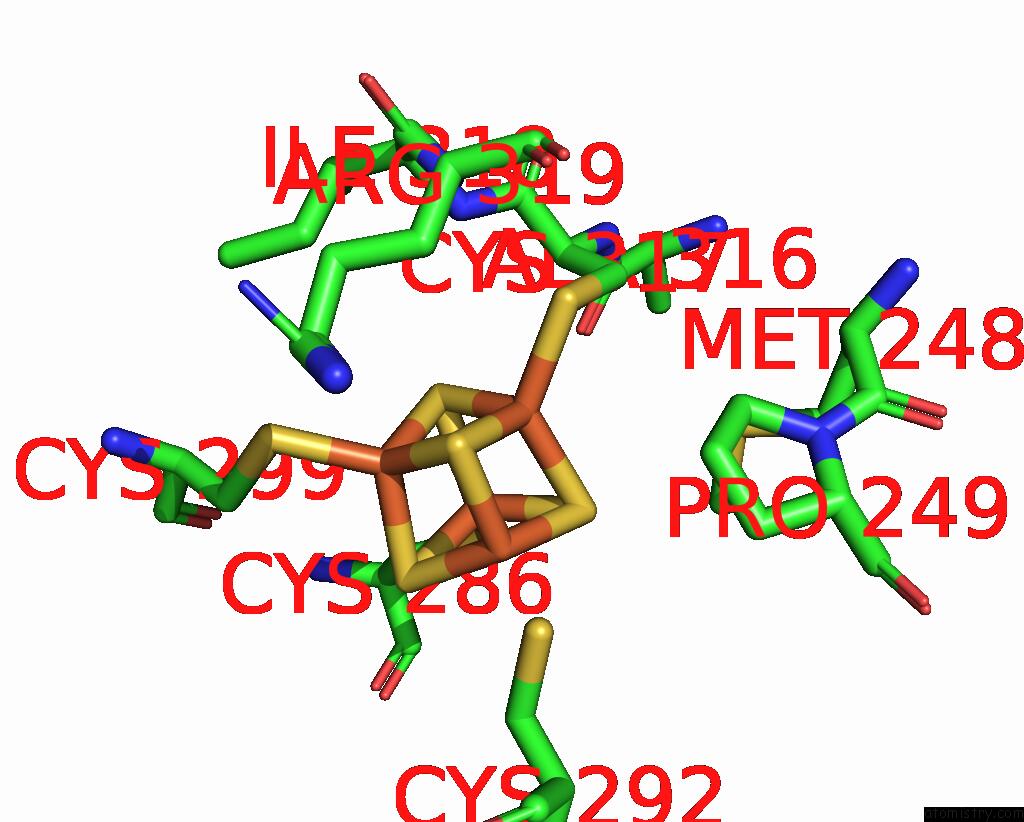
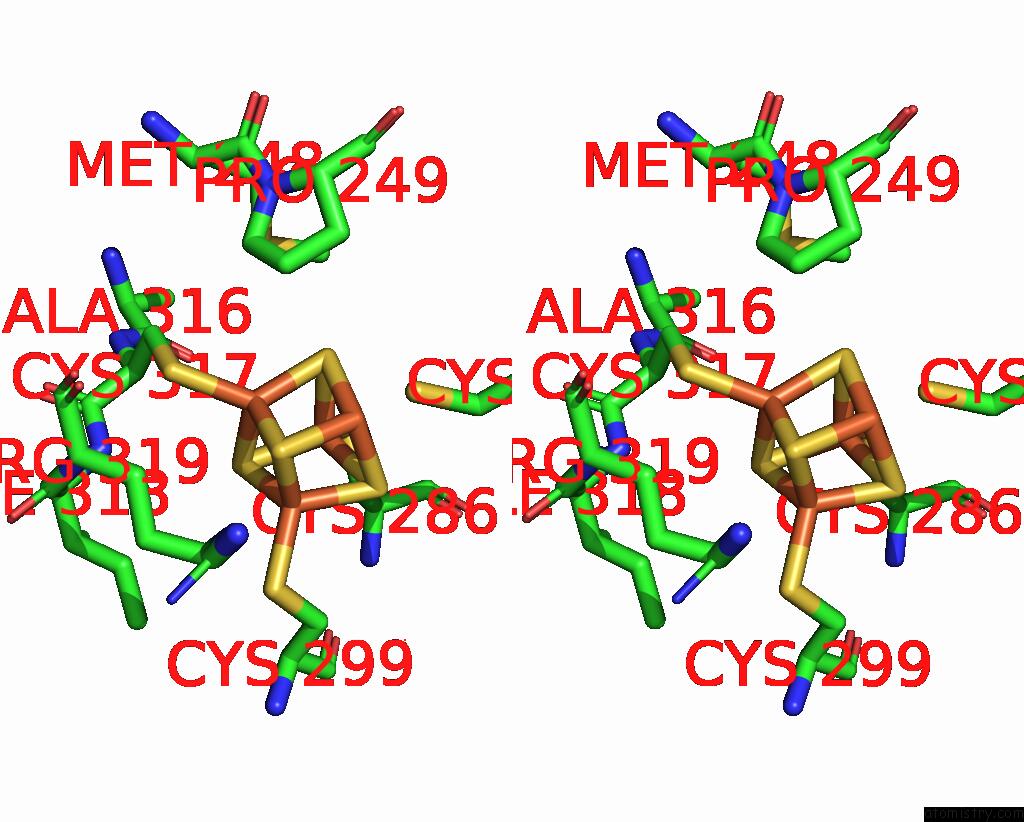
Iron binding site 4 out of 4 in 8j5r
Go back to
Iron binding site 4 out
of 4 in the Cryo-Em Structure of Mycobacterium Tuberculosis Oppabcd in the Resting State
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 4 of Cryo-Em Structure of Mycobacterium Tuberculosis Oppabcd in the Resting State within 5.0Å range:
|
Reference:
X.Yang,
T.Hu,
J.Liang,
Z.Xiong,
Z.Lin,
Y.Zhao,
X.Zhou,
Y.Gao,
S.Sun,
X.Yang,
L.W.Guddat,
H.Yang,
Z.Rao,
B.Zhang.
An Oligopeptide Permease, Oppabcd, Requires An Iron-Sulfur Cluster Domain For Functionality Nat.Struct.Mol.Biol. 2024.
ISSN: ESSN 1545-9985
DOI: 10.1038/S41594-024-01256-Z
Page generated: Sat Aug 10 06:31:25 2024
ISSN: ESSN 1545-9985
DOI: 10.1038/S41594-024-01256-Z
Last articles
Zn in 9J0NZn in 9J0O
Zn in 9J0P
Zn in 9FJX
Zn in 9EKB
Zn in 9C0F
Zn in 9CAH
Zn in 9CH0
Zn in 9CH3
Zn in 9CH1