Iron »
PDB 8ire-8jfw »
8jej »
Iron in PDB 8jej: Cryo-Em Structure of Na-Dithionite Reduced Membrane-Bound Fructose Dehydrogenase From Gluconobacter Japonicus
Enzymatic activity of Cryo-Em Structure of Na-Dithionite Reduced Membrane-Bound Fructose Dehydrogenase From Gluconobacter Japonicus
All present enzymatic activity of Cryo-Em Structure of Na-Dithionite Reduced Membrane-Bound Fructose Dehydrogenase From Gluconobacter Japonicus:
1.1.99.11;
1.1.99.11;
Iron Binding Sites:
The binding sites of Iron atom in the Cryo-Em Structure of Na-Dithionite Reduced Membrane-Bound Fructose Dehydrogenase From Gluconobacter Japonicus
(pdb code 8jej). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 6 binding sites of Iron where determined in the Cryo-Em Structure of Na-Dithionite Reduced Membrane-Bound Fructose Dehydrogenase From Gluconobacter Japonicus, PDB code: 8jej:
Jump to Iron binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Iron where determined in the Cryo-Em Structure of Na-Dithionite Reduced Membrane-Bound Fructose Dehydrogenase From Gluconobacter Japonicus, PDB code: 8jej:
Jump to Iron binding site number: 1; 2; 3; 4; 5; 6;
Iron binding site 1 out of 6 in 8jej
Go back to
Iron binding site 1 out
of 6 in the Cryo-Em Structure of Na-Dithionite Reduced Membrane-Bound Fructose Dehydrogenase From Gluconobacter Japonicus
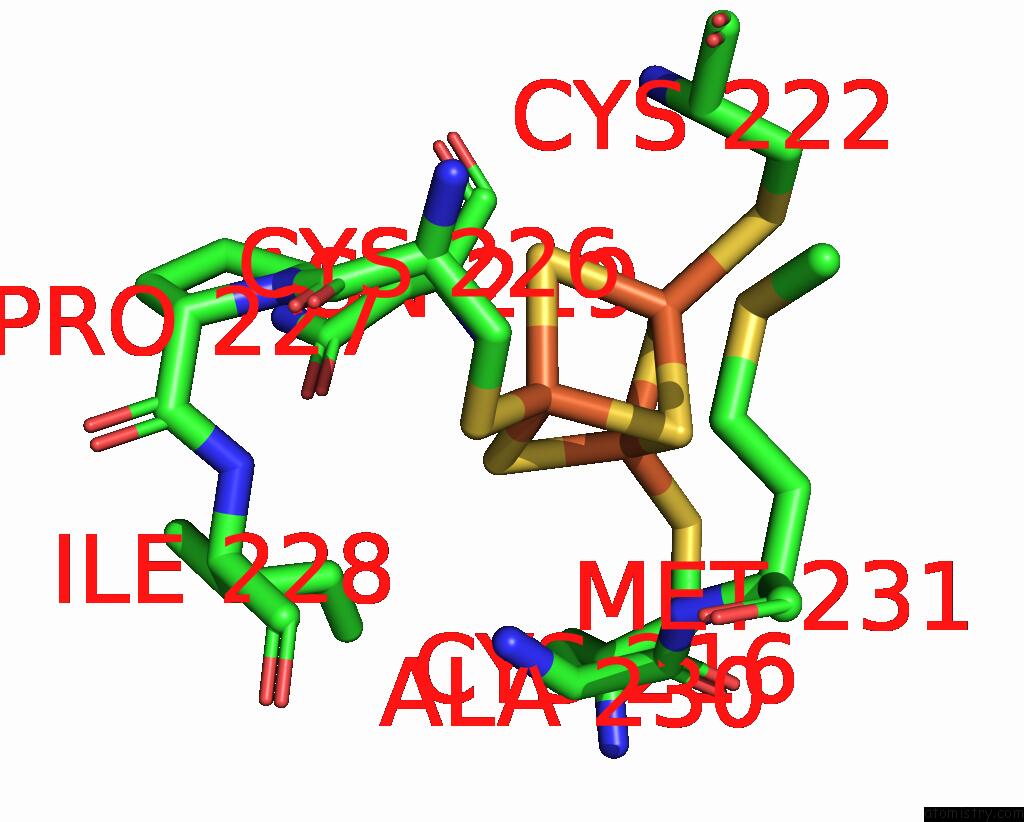
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Cryo-Em Structure of Na-Dithionite Reduced Membrane-Bound Fructose Dehydrogenase From Gluconobacter Japonicus within 5.0Å range:
|
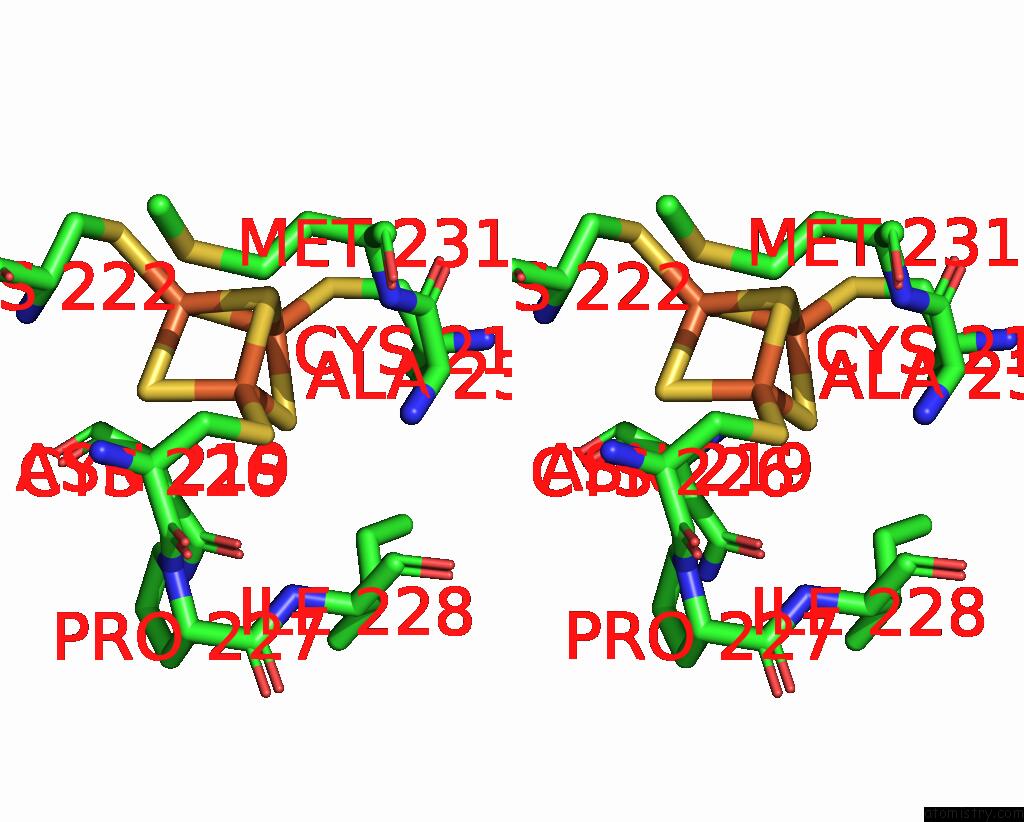
Iron binding site 2 out of 6 in 8jej
Go back to
Iron binding site 2 out
of 6 in the Cryo-Em Structure of Na-Dithionite Reduced Membrane-Bound Fructose Dehydrogenase From Gluconobacter Japonicus

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Cryo-Em Structure of Na-Dithionite Reduced Membrane-Bound Fructose Dehydrogenase From Gluconobacter Japonicus within 5.0Å range:
|
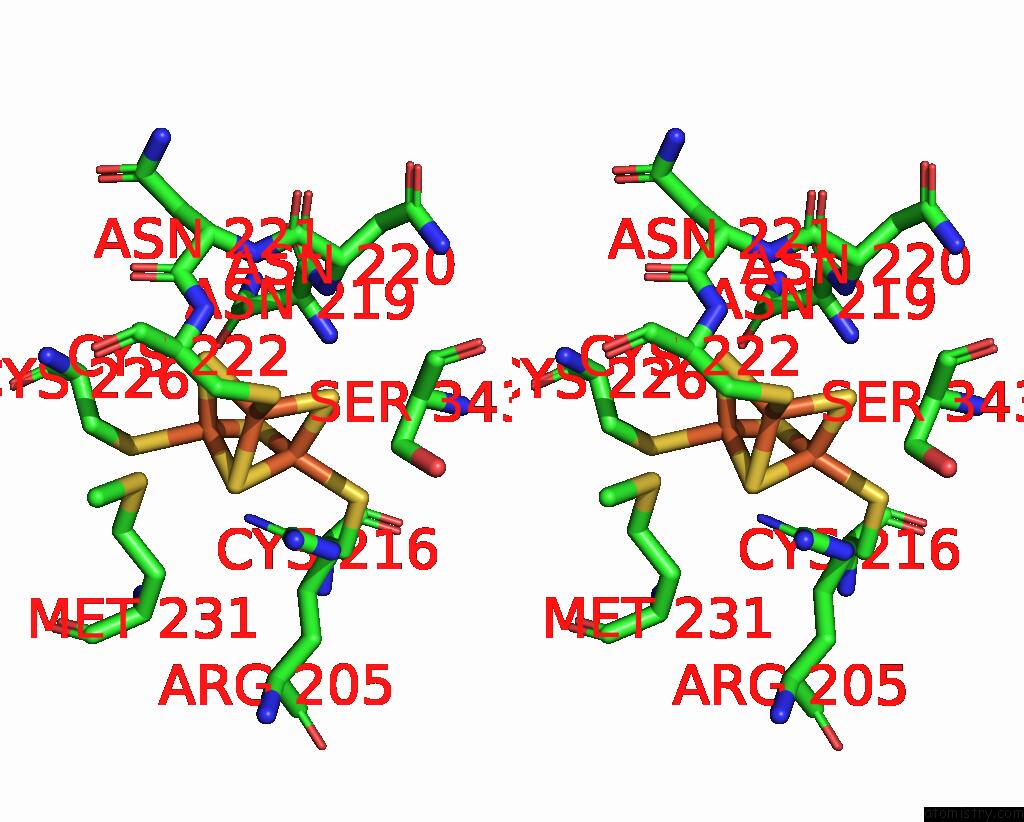
Iron binding site 3 out of 6 in 8jej
Go back to
Iron binding site 3 out
of 6 in the Cryo-Em Structure of Na-Dithionite Reduced Membrane-Bound Fructose Dehydrogenase From Gluconobacter Japonicus

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 3 of Cryo-Em Structure of Na-Dithionite Reduced Membrane-Bound Fructose Dehydrogenase From Gluconobacter Japonicus within 5.0Å range:
|
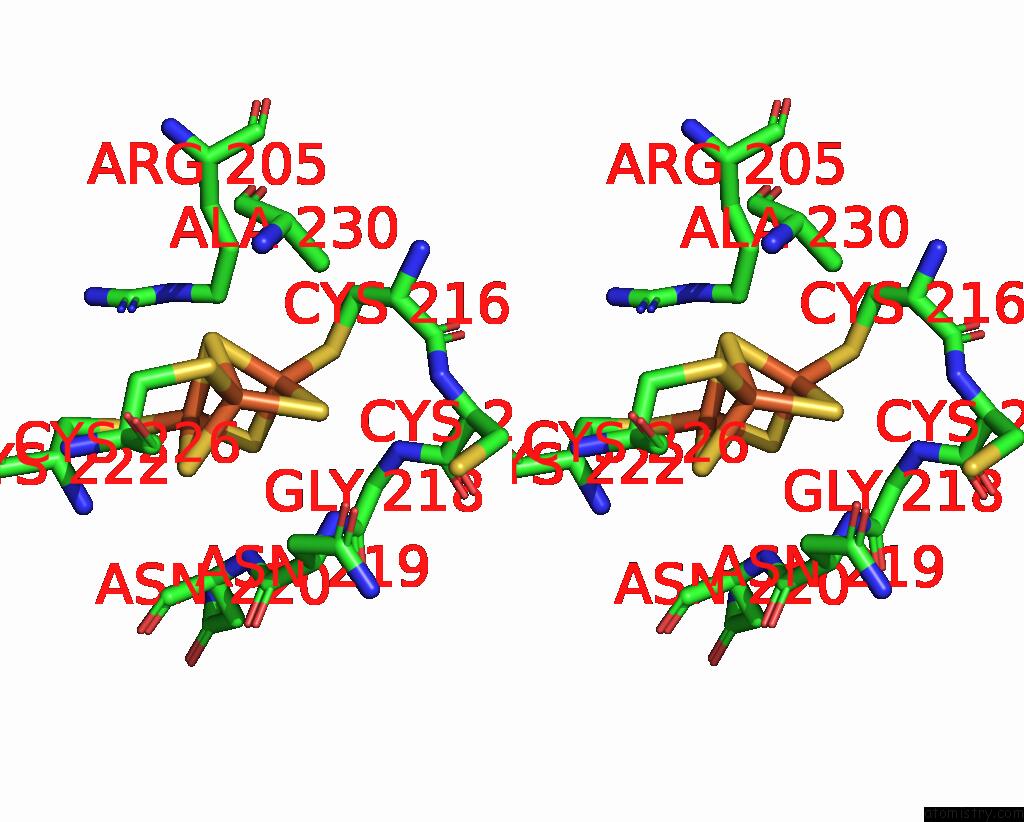
Iron binding site 4 out of 6 in 8jej
Go back to
Iron binding site 4 out
of 6 in the Cryo-Em Structure of Na-Dithionite Reduced Membrane-Bound Fructose Dehydrogenase From Gluconobacter Japonicus
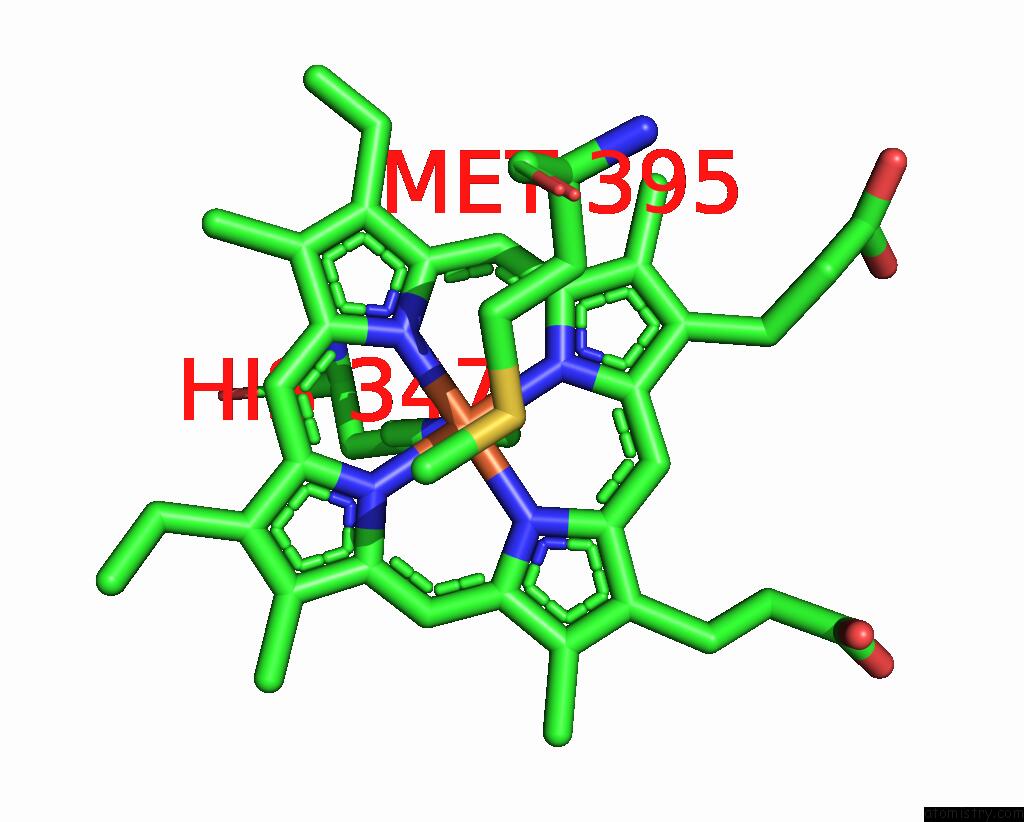
Mono view
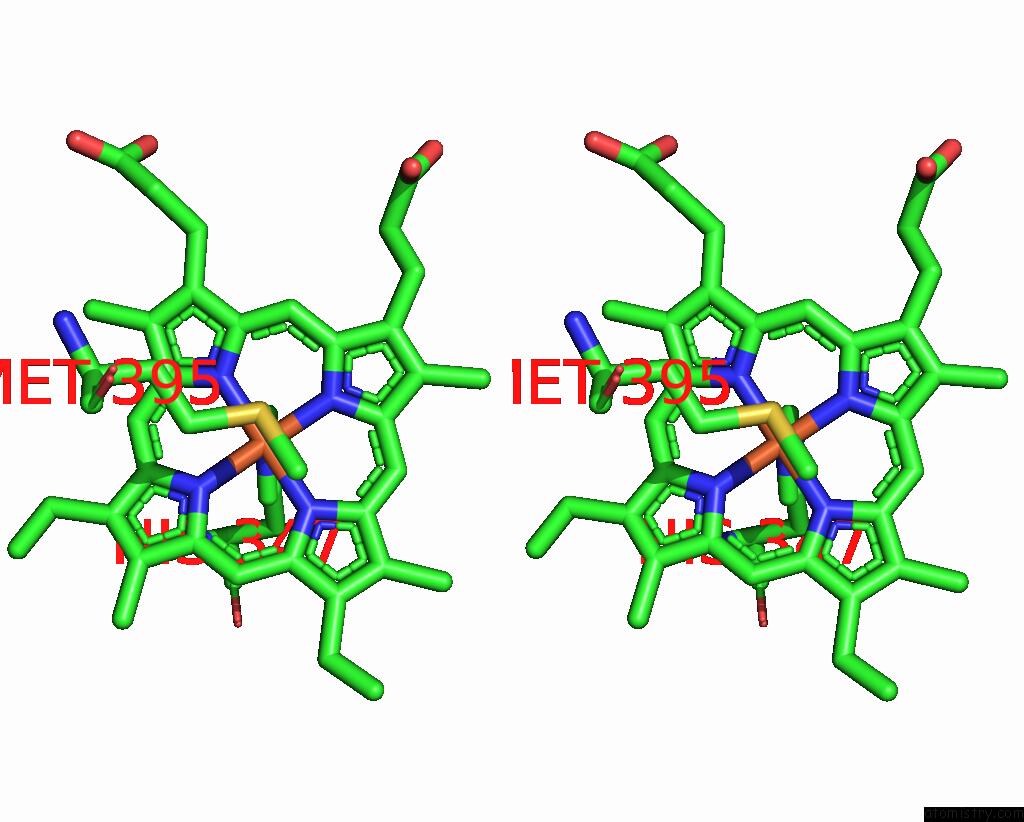
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 4 of Cryo-Em Structure of Na-Dithionite Reduced Membrane-Bound Fructose Dehydrogenase From Gluconobacter Japonicus within 5.0Å range:
|
Iron binding site 5 out of 6 in 8jej
Go back to
Iron binding site 5 out
of 6 in the Cryo-Em Structure of Na-Dithionite Reduced Membrane-Bound Fructose Dehydrogenase From Gluconobacter Japonicus
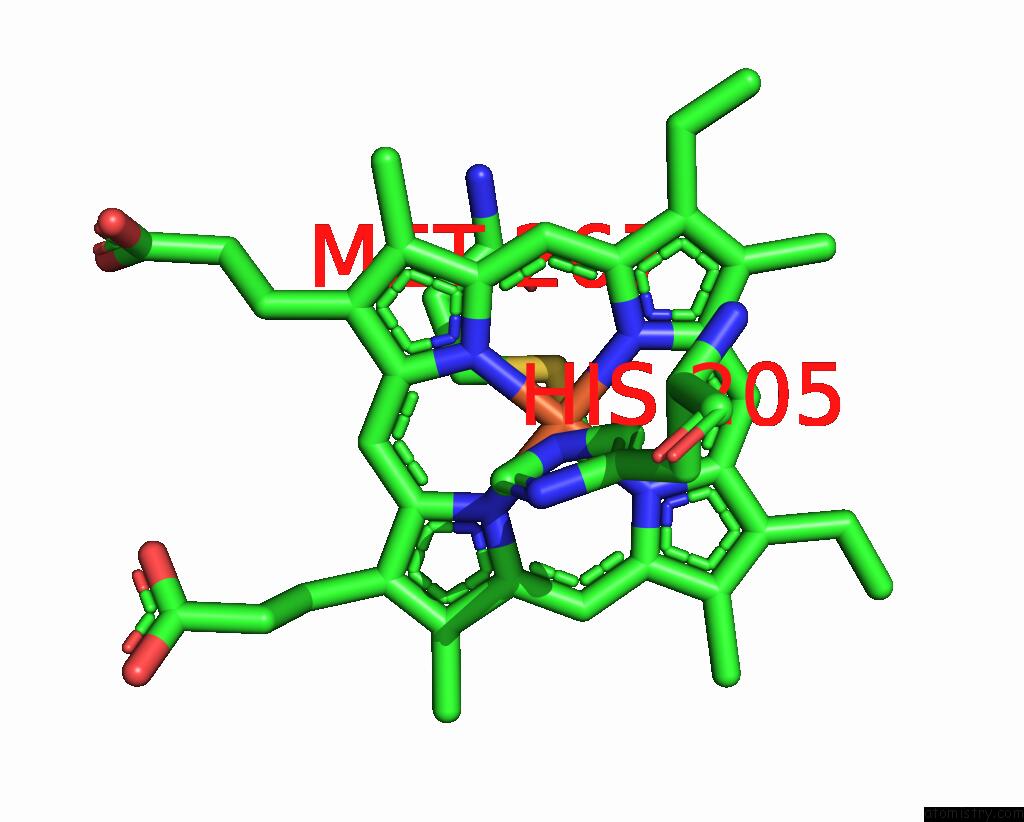
Mono view
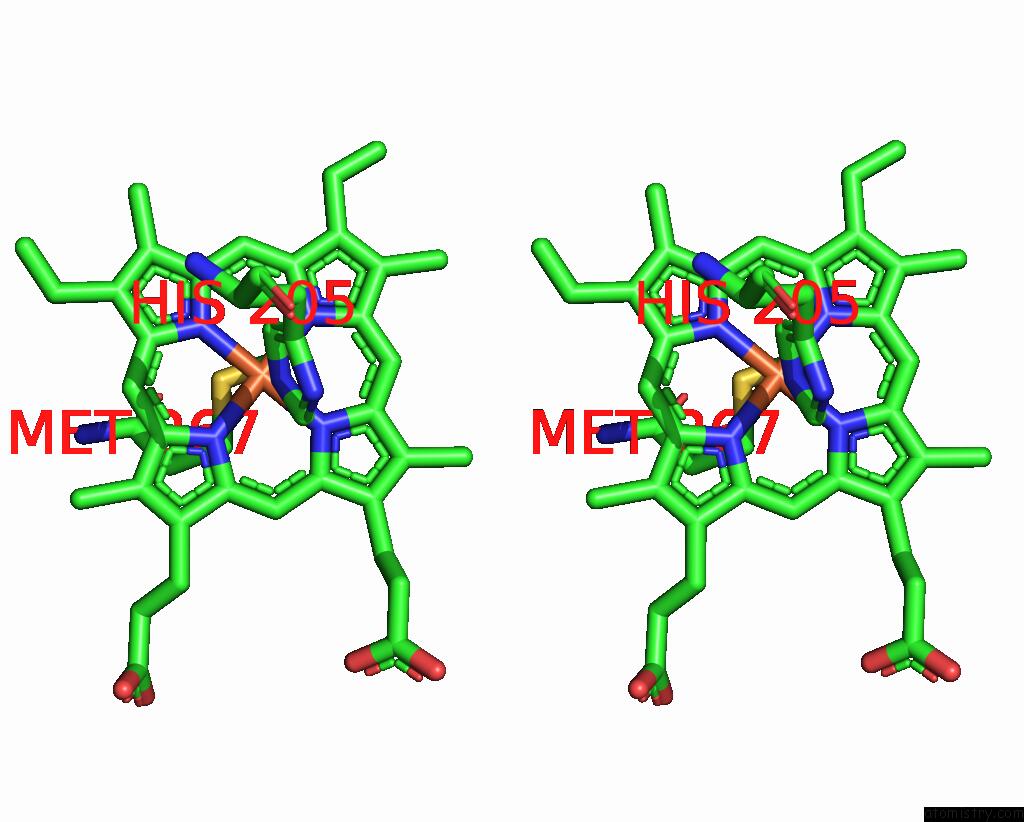
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 5 of Cryo-Em Structure of Na-Dithionite Reduced Membrane-Bound Fructose Dehydrogenase From Gluconobacter Japonicus within 5.0Å range:
|
Iron binding site 6 out of 6 in 8jej
Go back to
Iron binding site 6 out
of 6 in the Cryo-Em Structure of Na-Dithionite Reduced Membrane-Bound Fructose Dehydrogenase From Gluconobacter Japonicus
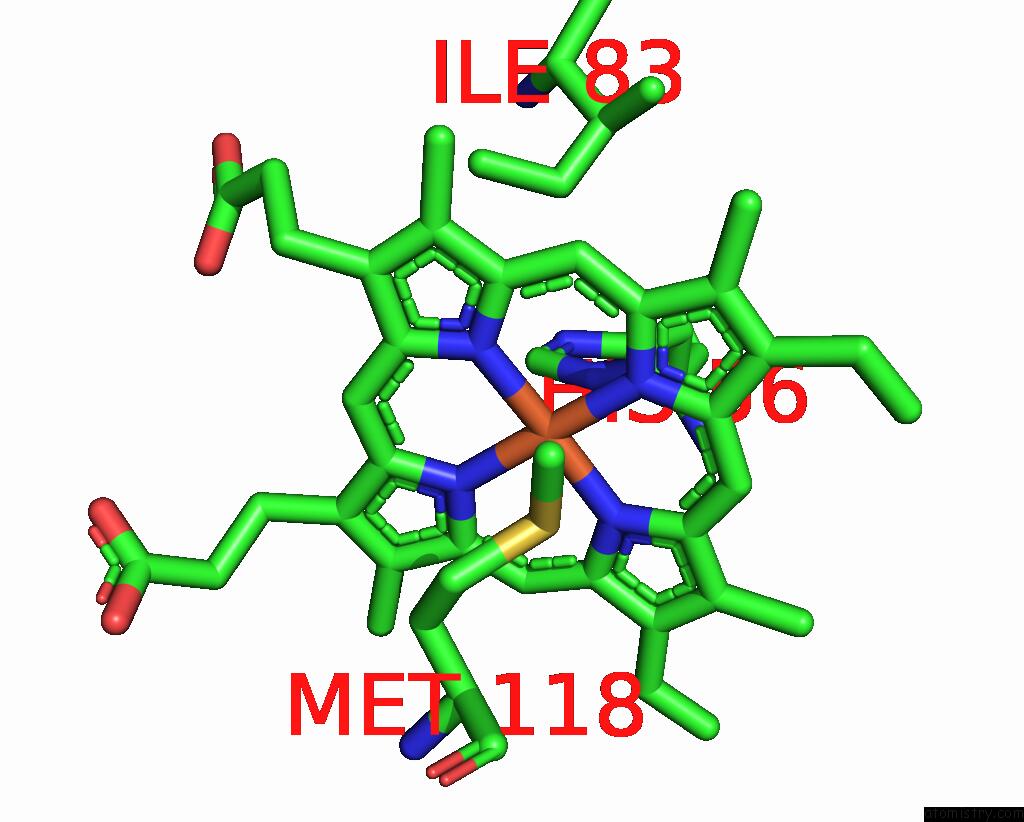
Mono view
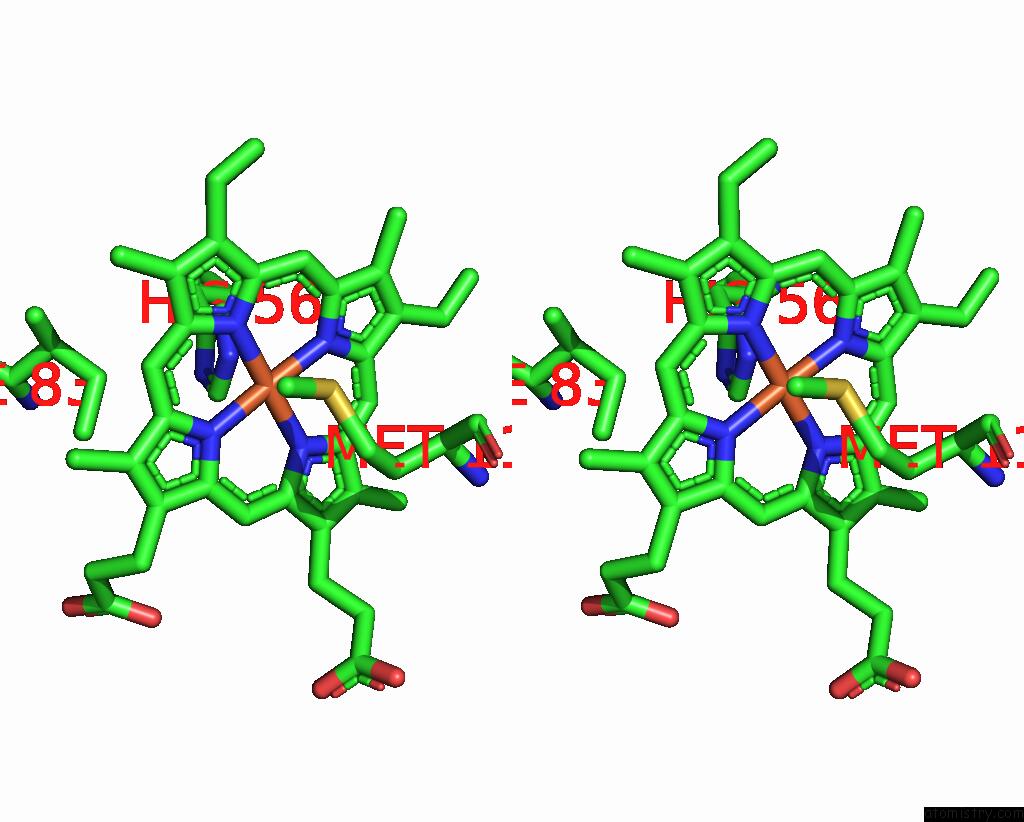
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 6 of Cryo-Em Structure of Na-Dithionite Reduced Membrane-Bound Fructose Dehydrogenase From Gluconobacter Japonicus within 5.0Å range:
|
Reference:
Y.Suzuki,
F.Makino,
T.Miyata,
H.Tanaka,
K.Namba,
K.Kano,
K.Sowa,
Y.Kitazumi,
O.Shirai.
Essential Insight of Direct Electron Transfer-Type Bioelectrocatalysis By Membrane-Bound D-Fructose Dehydrogenase with Structural Bioelectrochemistry. Acs Catalysis V. 13 2023.
ISSN: ESSN 2155-5435
DOI: 10.1021/ACSCATAL.3C03769
Page generated: Sat Aug 10 06:49:23 2024
ISSN: ESSN 2155-5435
DOI: 10.1021/ACSCATAL.3C03769
Last articles
Fe in 2YXOFe in 2YRS
Fe in 2YXC
Fe in 2YNM
Fe in 2YVJ
Fe in 2YP1
Fe in 2YU2
Fe in 2YU1
Fe in 2YQB
Fe in 2YOO