Iron »
PDB 8qbz-8qwt »
8qdr »
Iron in PDB 8qdr: Vitis Vinifera Dimeric 13S-Lipoxygenase Loxa in A Detergent Bound Open Conformation
Protein crystallography data
The structure of Vitis Vinifera Dimeric 13S-Lipoxygenase Loxa in A Detergent Bound Open Conformation, PDB code: 8qdr
was solved by
K.Wild,
I.Sinning,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 40.80 / 2.00 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 85.905, 105.818, 88.533, 90, 92.84, 90 |
| R / Rfree (%) | 17.4 / 21.8 |
Iron Binding Sites:
The binding sites of Iron atom in the Vitis Vinifera Dimeric 13S-Lipoxygenase Loxa in A Detergent Bound Open Conformation
(pdb code 8qdr). This binding sites where shown within
5.0 Angstroms radius around Iron atom.
In total 2 binding sites of Iron where determined in the Vitis Vinifera Dimeric 13S-Lipoxygenase Loxa in A Detergent Bound Open Conformation, PDB code: 8qdr:
Jump to Iron binding site number: 1; 2;
In total 2 binding sites of Iron where determined in the Vitis Vinifera Dimeric 13S-Lipoxygenase Loxa in A Detergent Bound Open Conformation, PDB code: 8qdr:
Jump to Iron binding site number: 1; 2;
Iron binding site 1 out of 2 in 8qdr
Go back to
Iron binding site 1 out
of 2 in the Vitis Vinifera Dimeric 13S-Lipoxygenase Loxa in A Detergent Bound Open Conformation
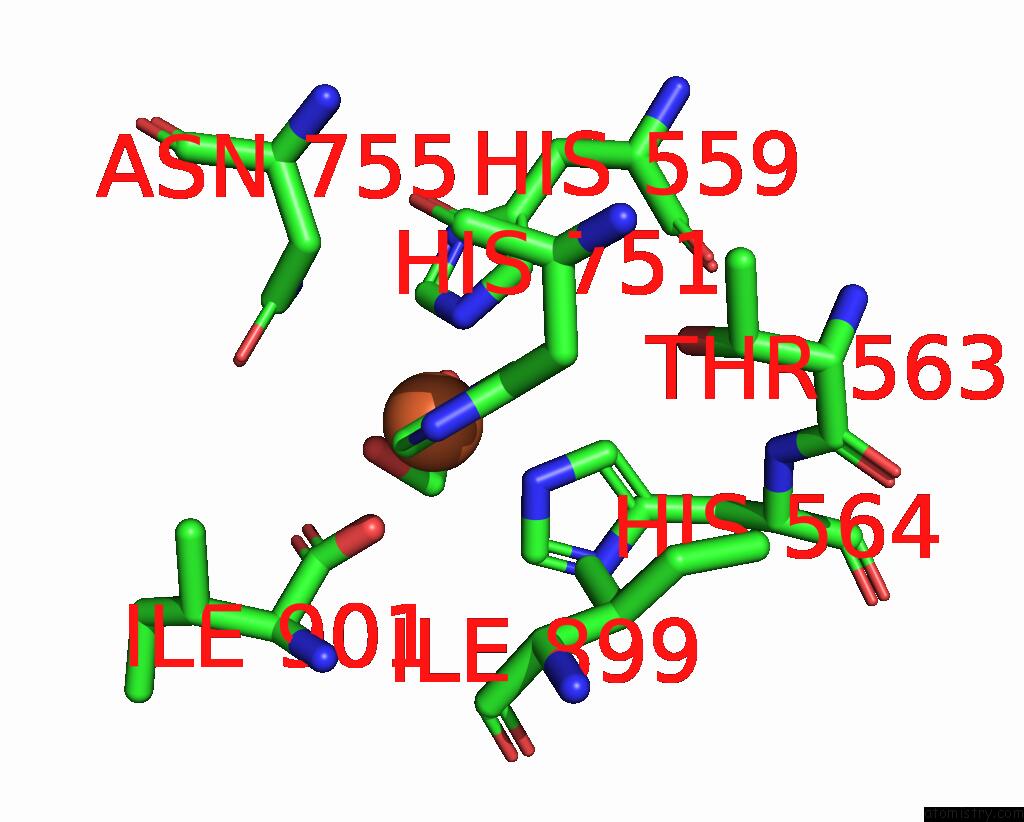
Mono view
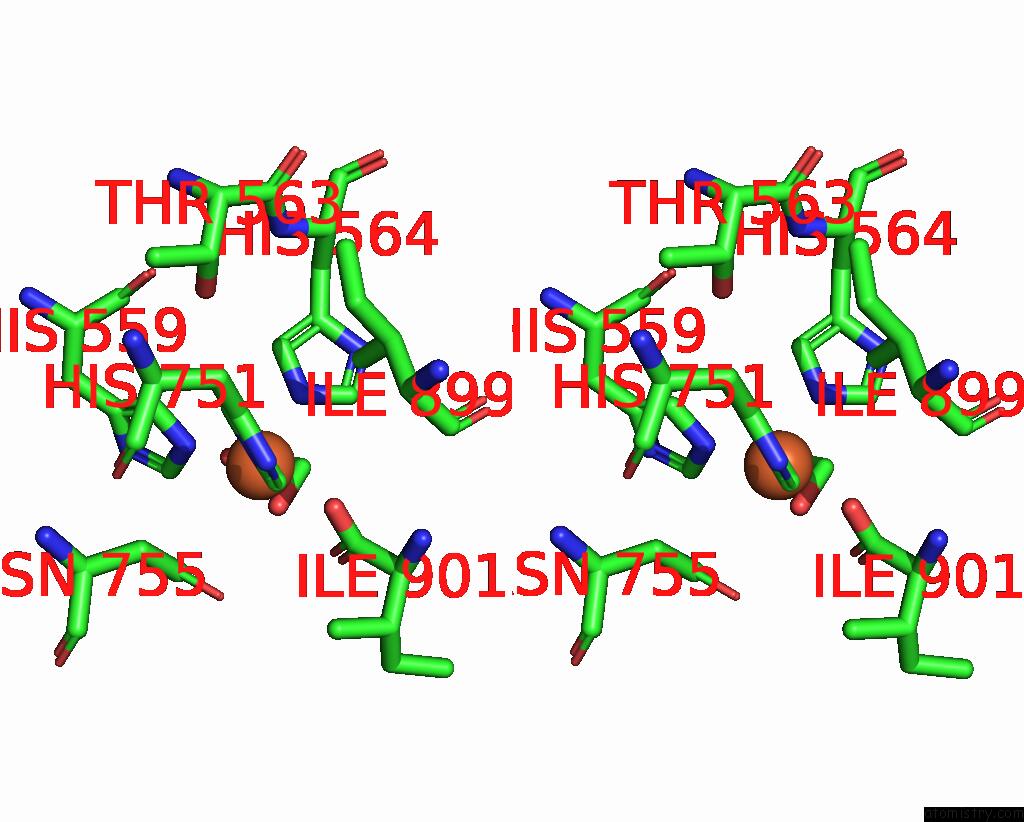
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 1 of Vitis Vinifera Dimeric 13S-Lipoxygenase Loxa in A Detergent Bound Open Conformation within 5.0Å range:
|
Iron binding site 2 out of 2 in 8qdr
Go back to
Iron binding site 2 out
of 2 in the Vitis Vinifera Dimeric 13S-Lipoxygenase Loxa in A Detergent Bound Open Conformation
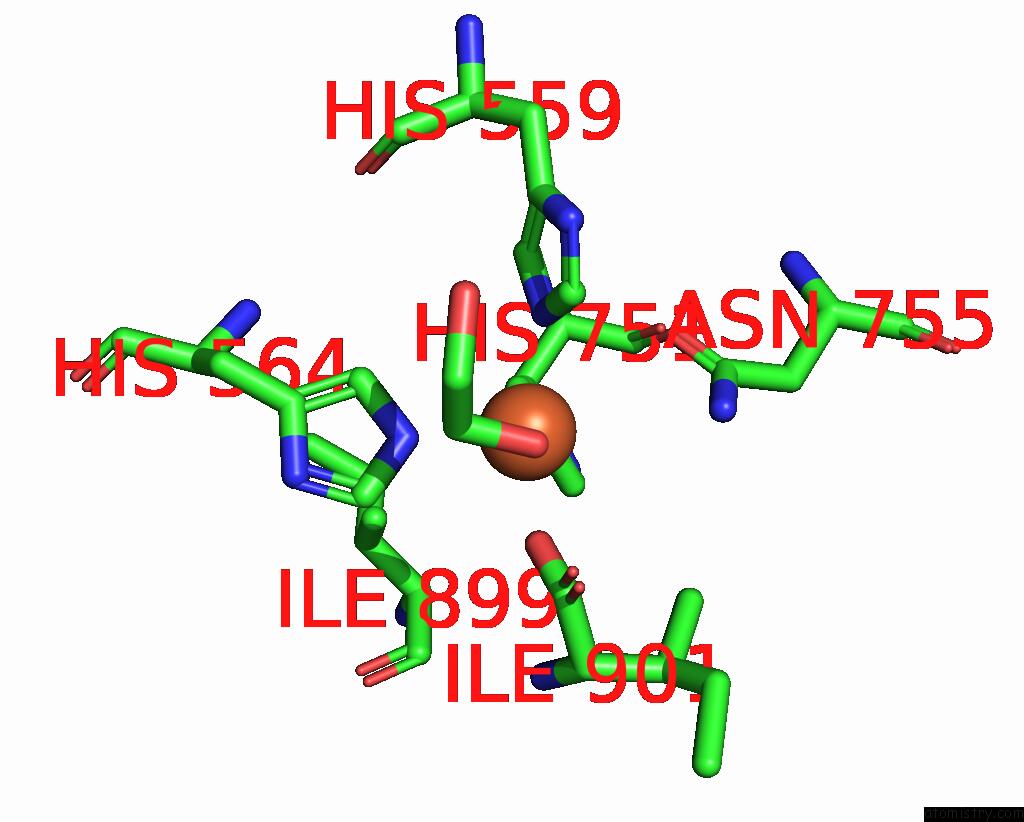
Mono view
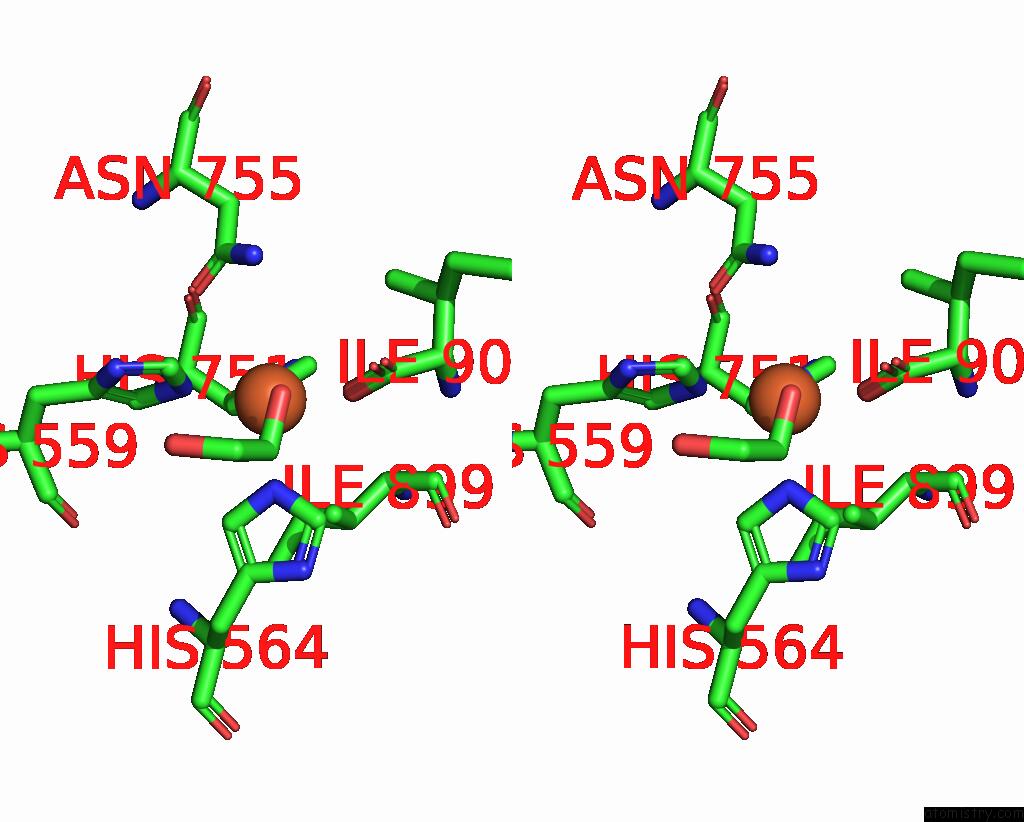
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iron with other atoms in the Fe binding
site number 2 of Vitis Vinifera Dimeric 13S-Lipoxygenase Loxa in A Detergent Bound Open Conformation within 5.0Å range:
|
Reference:
S.Pilati,
K.Wild,
A.Gumiero,
I.Holdermann,
Y.Hackmann,
M.Dalla Serra,
G.Guella,
C.Moser,
I.Sinning.
Vitis Vinifera Lipoxygenase Loxa Is An Allosteric Dimer Activated By Lipidic Surfaces. J.Mol.Biol. 68821 2024.
ISSN: ESSN 1089-8638
PubMed: 39424098
DOI: 10.1016/J.JMB.2024.168821
Page generated: Wed Nov 13 09:48:23 2024
ISSN: ESSN 1089-8638
PubMed: 39424098
DOI: 10.1016/J.JMB.2024.168821
Last articles
Zn in 9JYWZn in 9IR4
Zn in 9IR3
Zn in 9GMX
Zn in 9GMW
Zn in 9JEJ
Zn in 9ERF
Zn in 9ERE
Zn in 9EGV
Zn in 9EGW